 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000050G0250 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 610aa MW: 67579.9 Da PI: 5.9233 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 436.7 | 2.2e-133 | 38 | 413 | 1 | 351 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraW 94
eel +rmwkd+++l+r+ker+ +l ++a++++++ +++++qa rkkmsraQDgiLkYMlk mevcna+GfvYgiip+kgkpv+gasd++raW
PH01000050G0250 38 EELARRMWKDRVRLRRIKERQHKLA-LQQAELEKSRPKQISDQALRKKMSRAQDGILKYMLKLMEVCNASGFVYGIIPDKGKPVSGASDNIRAW 130
8********************9865.66677*************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--T CS
EIN3 95 WkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglsk 188
Wkekv+fd+ngpaai+ky+a+nl++++++s+ + +++hsl++lqD+tlgSLLs+lmqhc+ppqr++plekg++pPWWP+G+e+ww +lgl++
PH01000050G0250 131 WKEKVKFDKNGPAAIAKYEADNLVAANAQSS---GVKNQHSLMDLQDATLGSLLSSLMQHCNPPQRKYPLEKGTPPPWWPSGNEEWWIALGLPR 221
**************************88888...89********************************************************** PP
T--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX.XX....XXXXXXXXXXXX CS
EIN3 189 dqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah.ss....slrkqspkvtls 277
q ppykkphdlkk+wkv+vLt vikhmsp++++ir+++r+sk+lqdkm+akes ++l vl++ee++++++ s+ s ++++ t+s
PH01000050G0250 222 GQI-PPYKKPHDLKKVWKVGVLTGVIKHMSPNFDKIRNHVRKSKCLQDKMTAKESLIWLGVLQREERLVHSIDNGvSEithhSAPEDRNGDTHS 314
**9.9****************************************************************9999996446655555689999*** PP
XXXXXXXXXXXXXX.XXXXXXXXXX................................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XX CS
EIN3 278 ceqkedvegkkeskikhvqavktta................................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgset 339
+++++dv+g +e +++++++++ g++ krk+ +++s+++ ++ + + +++
PH01000050G0250 315 SSNEYDVDGFEEAPLSTSSKDDEHGlspvaqssdehgprrremantkcpnqvvpikaGTKGPPKRKRARHSSTAIVVQR------TDDTP-ENS 401
********99999989999988877888888888888888888888888875555555555555554444444443332......23333.345 PP
XXXXXXXXXXXX CS
EIN3 340 elifadknsisq 351
++ ++d+n+++q
PH01000050G0250 402 RNLIPDMNRLDQ 413
556666666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.3E-127 | 38 | 286 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 4.9E-70 | 160 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.57E-60 | 166 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 610 aa Download sequence Send to blast |
MEHLAILATE LVDSSDFEVD GIQNLTENDV SDEEIEPEEL ARRMWKDRVR LRRIKERQHK 60 LALQQAELEK SRPKQISDQA LRKKMSRAQD GILKYMLKLM EVCNASGFVY GIIPDKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIAKYEA DNLVAANAQS SGVKNQHSLM DLQDATLGSL 180 LSSLMQHCNP PQRKYPLEKG TPPPWWPSGN EEWWIALGLP RGQIPPYKKP HDLKKVWKVG 240 VLTGVIKHMS PNFDKIRNHV RKSKCLQDKM TAKESLIWLG VLQREERLVH SIDNGVSEIT 300 HHSAPEDRNG DTHSSSNEYD VDGFEEAPLS TSSKDDEHGL SPVAQSSDEH GPRRREMANT 360 KCPNQVVPIK AGTKGPPKRK RARHSSTAIV VQRTDDTPEN SRNLIPDMNR LDQVEIPGMA 420 NQITSFSQGD NTSEALQHRG DAQGQVHLPD AGVNNFDSAQ AANATPVSIY MGGQPVPYEG 480 CDNVRSKSGN TFPLDADSGF NNLPSSYQAL PLKQSLPLSM MDHHVVPMGI RAPADNSPYG 540 DPVIGSGIST SVPGDMQQLI DYPFYGEQDK FVGSSFEGLP LDYISISSPI PDIDDLLLHD 600 DDLMEYLGT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 4e-66 | 166 | 290 | 9 | 133 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
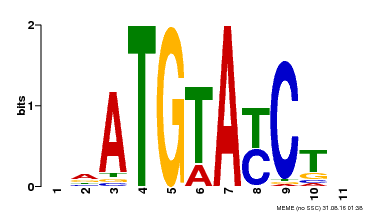
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000050G0250 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006660770.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_015696822.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-146 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A0A9DKK8 | 0.0 | A0A0A9DKK8_ARUDO; Uncharacterized protein | ||||
| STRING | OB09G21280.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-148 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



