 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000086G0840 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 294aa MW: 31539.4 Da PI: 8.5989 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 203.3 | 7.1e-63 | 1 | 139 | 6 | 145 |
DUF822 6 kptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl.eeaeaagssasaspesslq.ss 97
+pt kErEnnkrRERrRRaiaaki++GLRa G yklpk++DnneVlkALcreAGwvvedDGttyrkg+kp + + + +sa +sp+ss+q +s
PH01000086G0840 1 MPTSKERENNKRRERRRRAIAAKIFTGLRAFGSYKLPKHCDNNEVLKALCREAGWVVEDDGTTYRKGCKPRpSGPVGGSASAGMSPCSSSQlLS 94
69*********************************************************************566666667778********9** PP
DUF822 98 lkssalaspvesysaspksssfpspssldsislasaasllpvlsvlsl 145
+ ss+++spv+sy+asp+sssfpsp++ld+ s ++llp+l+ l++
PH01000086G0840 95 APSSSFPSPVPSYHASPASSSFPSPTRLDNPSP---SCLLPFLRGLPN 139
****************************99844...678888776655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.2E-59 | 1 | 124 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 294 aa Download sequence Send to blast |
MPTSKERENN KRRERRRRAI AAKIFTGLRA FGSYKLPKHC DNNEVLKALC REAGWVVEDD 60 GTTYRKGCKP RPSGPVGGSA SAGMSPCSSS QLLSAPSSSF PSPVPSYHAS PASSSFPSPT 120 RLDNPSPSCL LPFLRGLPNL PPLRVSSSAP VTPPLSSPTA SRPPKVQKPD WDVDPFRHPF 180 FAVSAPASPT RGRRLEHPDT IPECDESDVS TVDSGRWISF QMATTAPASP TYNLVNAGAS 240 NSNSMEIEGM AGERGRGGPE FEFDKGRVTP WEGERIHEVA AEELELTLGV GAK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 8e-26 | 19 | 81 | 391 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 8e-26 | 19 | 81 | 391 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 8e-26 | 19 | 81 | 391 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 8e-26 | 19 | 81 | 391 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Mediates downstream brassinosteroid-regulated growth response and feedback inhibition of brassinosteroid biosynthetic genes. May act as transcriptional repressor by binding the brassinosteroid-response element (5'-CGTGCG-3') in the promoter of GRAS32 (AC Q9LWU9), another positive regulator of brassinosteroid signaling (By similarity). {ECO:0000250}. | |||||
| UniProt | Positive brassinosteroid-signaling protein. Mediates downstream brassinosteroid-regulated growth response and feedback inhibition of brassinosteroid (BR) biosynthetic genes (PubMed:17699623, PubMed:19220793). May act as transcriptional repressor by binding the brassinosteroid-response element (BREE) (5'-CGTG(T/C)G-3') in the promoter of DLT (AC Q9LWU9), another positive regulator of BR signaling (PubMed:19220793). Acts as transcriptional repressor of LIC, a negative regulator of BR signaling, by binding to the BRRE element of its promoter. BZR1 and LIC play opposite roles in BR signaling and regulation of leaf bending (PubMed:22570626). {ECO:0000269|PubMed:17699623, ECO:0000269|PubMed:19220793, ECO:0000269|PubMed:22570626}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00073 | ChIP-chip | Transfer from AT1G19350 | Download |
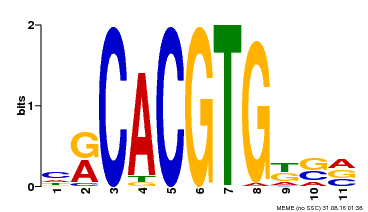
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000086G0840 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by 24-epibrassinolide. {ECO:0000269|PubMed:22570626}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK354370 | 0.0 | AK354370.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1006N24. | |||
| GenBank | AK359453 | 0.0 | AK359453.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1097E19. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020151437.1 | 1e-157 | protein BZR1 homolog 1 | ||||
| Swissprot | B8B7S5 | 1e-156 | BZR1_ORYSI; Protein BZR1 homolog 1 | ||||
| Swissprot | Q7XI96 | 1e-156 | BZR1_ORYSJ; Protein BZR1 homolog 1 | ||||
| TrEMBL | A0A446M3E3 | 1e-160 | A0A446M3E3_TRITD; Uncharacterized protein | ||||
| STRING | Traes_2BS_31BB97552.1 | 1e-157 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5474 | 34 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19350.6 | 2e-65 | BES1 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



