 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000129G0670 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 986aa MW: 110499 Da PI: 5.9902 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 174.8 | 1.1e-54 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
lke ++rwl+++ei++iL+n+++++++ e+++rp+sgsl+L++rk++ryfrkDG++w+kk+dgktv+E he+LK g+++vl+cyYah+een +f
PH01000129G0670 19 LKEaQHRWLRPTEICEILKNYRNFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKRDGKTVKEGHERLKSGSIDVLHCYYAHGEENINF 112
45559***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+yw+Lee++ +ivlvhylevk
PH01000129G0670 113 QRRSYWMLEEDFMHIVLVHYLEVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.929 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.0E-74 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.0E-48 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 2.3E-4 | 406 | 492 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 5.7E-4 | 407 | 484 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 5.0E-17 | 407 | 492 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 5.13E-17 | 588 | 693 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.0E-17 | 591 | 699 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.63E-13 | 595 | 689 | No hit | No description |
| PROSITE profile | PS50088 | 8.79 | 597 | 629 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 17.9 | 597 | 701 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.736 | 630 | 662 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.013 | 630 | 659 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1200 | 669 | 698 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.55E-8 | 802 | 854 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.075 | 803 | 825 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 804 | 833 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0032 | 805 | 824 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 826 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.578 | 827 | 851 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.9E-4 | 829 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 986 aa Download sequence Send to blast |
MAEGRRYAIA PQLDIEQILK EAQHRWLRPT EICEILKNYR NFRIAPEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKRD GKTVKEGHER LKSGSIDVLH CYYAHGEENI NFQRRSYWML 120 EEDFMHIVLV HYLEVKVGKS SSRSRVHDDM LQATHVDSPF SQLPSQTTEA ESSLSGQASE 180 YEETESGYHQ GLQATTLNTD FHSHSQDNLP VVLMESAPGI AFNGPNSQFD LSSRNEVMKL 240 DKGIHQMPPY QAPVPSEQSP FTEGPGIESF TFDEVYNNGP SIKDIGGAGT DGEPLWQLPS 300 VIGGPFATVD SFQQNDRSLE EAINYPLLKT QSSNLSDILR DSFKKSDSFT RWMSKELAEV 360 DDSQIKSSSG VYWNSEEADN IIEASSHDQL DQFTLGPVLA QDQLFSIVDF SPSWAYAGSK 420 TKVLVFGRFI KSDEVKRFKW SCMFGEVEVP AEILANGTLR CYSPSHEPGR VPFYVTCSNR 480 LACSEVREFE FRPSPHGATN KTYLQMRLDK LLSLGQDECQ VTLSNPTKEM VDLSKKISLL 540 MMNNDTWSEL LKLADGNELA TDDTDQFLEN CIKEKMHIWL LHRVGDGGKG PGVLDEEGQG 600 VLHLAAALGY DWAIRPTITA GVNINFRDAH GWTALHWAAF CGRERTVVAL IALGAAPGAL 660 TDPTPDFPLG RTPADLASAN GHKGISGFLA ESSLTSHVQT LNLKEAMWGN AAEISGLPGI 720 GDVTERSVSP LAGEGLQAGS MGDSLGAVQN ATQAAARIYQ VFRVQSFQRK QAVQYEDDNG 780 AISDERALSL LSVKPSKPGQ LDPLHAAATR IQNKFRGWKG RKEFLLIRQR IVKIQAHVRG 840 HQVRKHYRKI IWSVGIVEKV ILRWRRRGAG LRGFRSAEGA TESTNSSSSD VIQNKPAEDD 900 YDFLQEGRKQ TEERLQKALA RVKSMVQYPD ARDQYQRILT VVTKMQESQA MQEKMLEEST 960 DMDEGFLTSE FRELWDDDMP MPGYF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
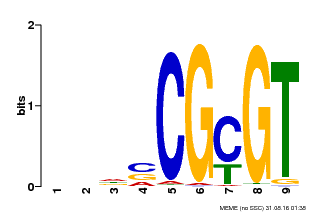
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000129G0670 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631249.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A0E0CWH5 | 0.0 | A0A0E0CWH5_9ORYZ; Uncharacterized protein | ||||
| STRING | Si034076m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP608 | 38 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



