 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000188G0440 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 245aa MW: 27129.5 Da PI: 6.4034 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.3 | 1.3e-18 | 31 | 84 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
PH01000188G0440 31 KKRRLNVEQVRTLEKNFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWK 84
4568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.1 | 3.7e-41 | 30 | 120 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqv++LE++Fe +kLeperK++lar+Lglqprqva+WFqnrRAR+ktkqlEk+y+aLkr++da+k+en++L +++++L++e+
PH01000188G0440 30 EKKRRLNVEQVRTLEKNFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWKTKQLEKEYDALKRQLDAVKAENDALLSHNKKLQAEI 120
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-19 | 12 | 87 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.94E-20 | 13 | 88 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.41 | 26 | 86 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-17 | 29 | 90 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.53E-17 | 31 | 87 | No hit | No description |
| Pfam | PF00046 | 6.1E-16 | 31 | 84 | IPR001356 | Homeobox domain |
| PRINTS | PR00031 | 6.9E-6 | 57 | 66 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 61 | 84 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 6.9E-6 | 66 | 82 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 7.7E-13 | 86 | 126 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 245 aa Download sequence Send to blast |
MAPMLGKQPM YGGGGANDDE LSDDGSQAGE KKRRLNVEQV RTLEKNFELG NKLEPERKMQ 60 LARALGLQPR QVAIWFQNRR ARWKTKQLEK EYDALKRQLD AVKAENDALL SHNKKLQAEI 120 LALKGREAAS ELINLNKETE ASCSNRSENS SEINLDISRT PPSESPMDPP PPHQQGGCMI 180 PFYPSVARTA GVDIDQLLHS SSVPKMEHGG ANVQASDTAS FGNLLCGVDE PPPFWPWADH 240 QHFH* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 78 | 86 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
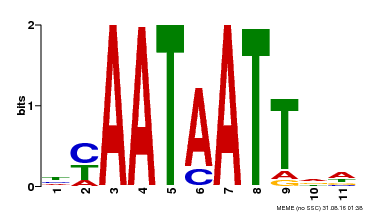
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000188G0440 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK359041 | 0.0 | AK359041.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1087P18. | |||
| GenBank | AK359747 | 0.0 | AK359747.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1102E15. | |||
| GenBank | AK366245 | 0.0 | AK366245.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2041G22. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004985543.1 | 1e-145 | homeobox-leucine zipper protein HOX21 isoform X2 | ||||
| Swissprot | Q8S7W9 | 1e-136 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | A0A3L6SDB5 | 1e-153 | A0A3L6SDB5_PANMI; Uncharacterized protein | ||||
| STRING | Pavir.Ia04323.1.p | 1e-153 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 1e-80 | HD-ZIP family protein | ||||



