 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000206G0250 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 561aa MW: 59322.4 Da PI: 6.1221 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.6 | 2.3e-32 | 223 | 280 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK++kgse+prsYY+C++agCp+kkkver d++v+ei+Y+g+Hnh+k
PH01000206G0250 223 DDGYNWRKYGQKQMKGSENPRSYYKCSFAGCPTKKKVERLP-DGQVTEIVYKGTHNHPK 280
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 103.3 | 1.3e-32 | 382 | 440 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+ gCpv+k ver+++d ++v++tYeg+Hnh+
PH01000206G0250 382 LDDGYRWRKYGQKVVKGNPNPRSYYKCTTVGCPVRKLVERASHDLRAVITTYEGKHNHD 440
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.4E-29 | 214 | 281 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.58E-25 | 218 | 281 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.6E-35 | 222 | 280 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.72 | 223 | 281 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.1E-25 | 223 | 279 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 4.1E-36 | 367 | 442 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.84E-29 | 374 | 442 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.447 | 377 | 442 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.6E-38 | 382 | 441 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.2E-25 | 383 | 440 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 561 aa Download sequence Send to blast |
MTSAPGSLGA LANSGPVALT FGTNSFANLM SGSASSGAAC GGTGVSKFKA MAPSSLPLSL 60 PPASPSACFS TPSGFLDSPI LLIPSLFPSP TTGAFPSQPF NWMGTPENLQ GGVKDEQRQY 120 SDFTFQTTMT GATTTAASSF LHSSVLMAPL GGDSYNGEQH QTWSYQEQSM DAATRPAEFA 180 APFEATSSGI TAVTAAPDML VNGGYNSMSG AGFREQQSRR PSDDGYNWRK YGQKQMKGSE 240 NPRSYYKCSF AGCPTKKKVE RLPDGQVTEI VYKGTHNHPK PQNARRGSGS APYALQGAND 300 ALSGTPVATP ENSSASYGDD EINGVSSPLV GNVGGGEEFD DDEPDSKKWR KDGDGEGISL 360 AGNRTVREPR VVVQTMSDID ILDDGYRWRK YGQKVVKGNP NPRSYYKCTT VGCPVRKLVE 420 RASHDLRAVI TTYEGKHNHD VPAARGSATL YRATPPAGKA CTRPTAPAAS YLHGSGGGYS 480 GDLRPDWFGA VQQGAFGGLD GGAPTQPADG GFALSGFDNP MGTSYSYTSQ QQQQNDAMYY 540 ASSTKDEPRD DIFFEQSLLF * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 3e-34 | 223 | 443 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 3e-34 | 223 | 443 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (By similarity). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
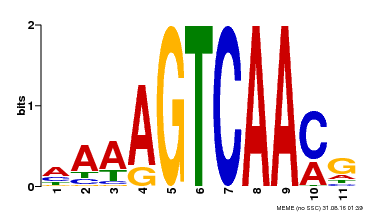
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000206G0250 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:25110688). Slightly down-regulated by gibberellic acid (GA) (By similarity). Accumulates in response to jasmonic acid (MeJA) (PubMed:16919842). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000269|PubMed:16919842, ECO:0000269|PubMed:25110688}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY676925 | 1e-131 | AY676925.1 Oryza sativa (indica cultivar-group) transcription factor WRKY24 (WRKY24) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025809961.1 | 0.0 | WRKY transcription factor WRKY24-like | ||||
| Swissprot | Q6B6R4 | 1e-166 | WRK24_ORYSI; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A3L6RDQ6 | 0.0 | A0A3L6RDQ6_PANMI; Putative WRKY transcription factor 25 | ||||
| STRING | Pavir.J03800.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2992 | 36 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 4e-94 | WRKY DNA-binding protein 33 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



