 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000241G1400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 371aa MW: 41031.9 Da PI: 10.2137 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 29.3 | 1.9e-09 | 5 | 34 | 1 | 30 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIart 30
+++WT eE+e l ++v ++G g W+tI +
PH01000241G1400 5 KQKWTSEEEEALRRGVLKHGAGKWRTIQKD 34
79*************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 9.989 | 1 | 58 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.16E-12 | 2 | 80 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.8E-4 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-10 | 5 | 51 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.5E-6 | 5 | 33 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 1.03E-14 | 6 | 83 | No hit | No description |
| SMART | SM00526 | 6.3E-9 | 187 | 252 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 20.754 | 189 | 257 | IPR005818 | Linker histone H1/H5, domain H15 |
| Pfam | PF00538 | 8.1E-8 | 193 | 249 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 9.8E-13 | 193 | 266 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 7.8E-11 | 193 | 263 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 371 aa Download sequence Send to blast |
MGAPKQKWTS EEEEALRRGV LKHGAGKWRT IQKDPEFSPV LSSRSNIDLK IQYMDKLVDS 60 SFPRLSKWEK LCIAGPVDKW RNLSFSASGL GSRDKIRVPK IKGPSSSPSP SSQALLLPAP 120 NKVAEALPPA DSEKSPQSQD GKTPKLASLY FHTEVQFIKL IRNMILANDC KMIFTPVGMT 180 MIKISDMKLR SWYSFMILEA LSELKDPNGS DVAAICNFIE QRHEVQPNFR RLLGAKLRRL 240 MGAKKIEKID KAYRLTDSYA TKASAPIKTP TPKQKDPAKP LKVKASQNLG LLSAVSPALD 300 AAAAAAVKVA DAEAKANLAN EHMIEAERIL KVAEQTESLL TLATEIYERC IPMSILQHFT 360 FHLPIIFSVI * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats, but may also bind to the single telomeric strand. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
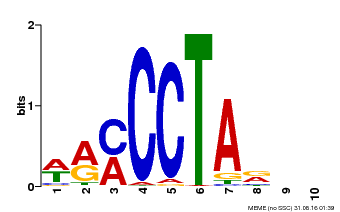
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000241G1400 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP094524 | 0.0 | FP094524.1 Phyllostachys edulis cDNA clone: bphylf024j07, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015622156.1 | 1e-116 | single myb histone 4 isoform X1 | ||||
| Swissprot | Q6WLH4 | 1e-106 | SMH3_MAIZE; Single myb histone 3 | ||||
| TrEMBL | A0A0A9PJS4 | 1e-121 | A0A0A9PJS4_ARUDO; Smh4 | ||||
| STRING | OPUNC01G28080.1 | 1e-116 | (Oryza punctata) | ||||
| STRING | ORGLA01G0233700.1 | 1e-116 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9247 | 34 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 2e-24 | telomere repeat binding factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



