 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000358G0250 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 290aa MW: 31209.4 Da PI: 10.0627 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 47.7 | 3.5e-15 | 112 | 156 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ +++ + ++lG+g+W+ I+r++ ++Rt+ q+ s+ qky
PH01000358G0250 112 PWTEDEHRKFLAGLEKLGKGDWRGISRHFVTTRTPTQVASHAQKY 156
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 20.049 | 105 | 161 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.92E-18 | 106 | 162 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.7E-18 | 108 | 160 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 6.0E-12 | 109 | 159 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-11 | 109 | 155 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.92E-10 | 112 | 157 | No hit | No description |
| Pfam | PF00249 | 2.1E-12 | 112 | 156 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 290 aa Download sequence Send to blast |
MARKCSICGN NGHNSRTCSG HRSLENSGGL RLFGVQLYAG SSSPLKKCFS MECLSSSASA 60 YYAAAVAASN SSPSVSSSSS LVSIDETTEK MTNGYLSDGL MGRAQERKKG VPWTEDEHRK 120 FLAGLEKLGK GDWRGISRHF VTTRTPTQVA SHAQKYFLRQ SSLTQKKRRS SLFDVVENGE 180 KATCVNERLR LKEETTPVSG MEFPTLSLGI SRPKTEAVLP PCLTLMPSCS SAVSNTSPNL 240 APKLPPALTA KPQVNLQTPD LELKISTSRQ ADHTGSSART PFFGTIRVS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
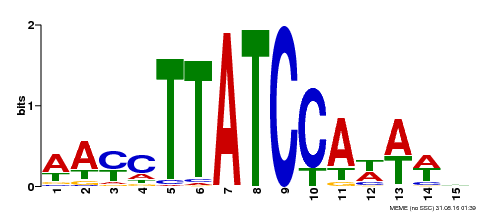
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000358G0250 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006643865.1 | 1e-116 | PREDICTED: transcription factor MYB1R1-like | ||||
| Refseq | XP_015625837.1 | 1e-116 | transcription factor MYBS3-like | ||||
| TrEMBL | A0A0A9ESZ5 | 1e-146 | A0A0A9ESZ5_ARUDO; Uncharacterized protein | ||||
| STRING | LPERR01G05630.1 | 1e-123 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4231 | 38 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 7e-50 | MYB_related family protein | ||||



