 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01001245G0180 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 592aa MW: 63226.2 Da PI: 6.2722 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102 | 3.6e-32 | 209 | 266 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkk +ers d++++e++Y+g+Hnh+k
PH01001245G0180 209 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCEVKKLLERSL-DGQITEVVYKGRHNHPK 266
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.5 | 1.4e-33 | 382 | 440 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct +gCpv+k+ver+++dpk v++tYeg+Hnhe
PH01001245G0180 382 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNTGCPVRKHVERASHDPKSVITTYEGKHNHE 440
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 6.6E-28 | 200 | 267 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.801 | 203 | 267 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.63E-25 | 207 | 267 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.6E-35 | 208 | 266 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.0E-24 | 209 | 265 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.0E-37 | 367 | 442 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-29 | 374 | 442 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.526 | 377 | 442 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.7E-39 | 382 | 441 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.3E-26 | 383 | 440 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 592 aa Download sequence Send to blast |
MADSPNPSSG DLPAGVGSSP ENPAPAYGRV AAGAGARYKA MSPARLPISR EPCLTIPAGF 60 SPSALLESPV LLTNFKVEPS PTTGTLSMAA IMNKSAHPNI LPSPRDNSVG TAPEDGGSRD 120 FEFKPHLNSS SQSLAPAIND QKNHEPSMQN QSLNPIVDVS SAPNQPVGMV GLTDSMPAEV 180 GPSDLHQMNS SENAVQETQS ENVAEKSAED GYNWRKYGQK HVKGSENPRS YYKCTHPNCE 240 VKKLLERSLD GQITEVVYKG RHNHPKPQPS RRLAAGAVPS NQGEERYDGV AATEDKSSNV 300 LSNLGNPVHS AGMVEPVPGS VSDDDIDAGG GRPYPGDDAT EDDDLESKRR KMESAGIDAA 360 LMGKPNREPR VVVQTVSEVD ILDDGYRWRK YGQKVVKGNP NPRSYYKCTN TGCPVRKHVE 420 RASHDPKSVI TTYEGKHNHE VPAARNASHE MSTPPMKTLV HPINSNMPGL GGMMRACEAR 480 TFTNQYSQAA ESDTISLDLG VGISPNHSDA TNQMRSSVPD QMQYQMQPMA SVYGSMGLPA 540 MPVPTVPGNA ANSIYGSREE KGSEGFTFKA TPMDHSANLC YSSAGNLVLG P* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-36 | 189 | 443 | 2 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-36 | 189 | 443 | 2 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
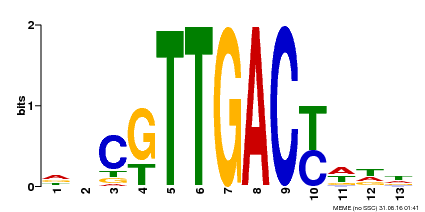
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01001245G0180 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM201384 | 0.0 | KM201384.1 Stipa purpurea WRKY17 transcription factor mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646742.1 | 0.0 | WRKY transcription factor SUSIBA2 isoform X1 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | A0A0B5GUL9 | 0.0 | A0A0B5GUL9_9POAL; WRKY17 transcription factor (Fragment) | ||||
| STRING | OMERI07G17160.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6740 | 38 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-125 | WRKY family protein | ||||



