 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01001316G0020 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 809aa MW: 88964.4 Da PI: 5.2503 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 110.1 | 1.4e-34 | 176 | 250 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv gCe+d+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
PH01001316G0020 176 CQVSGCEEDIRELKGYHRRHRVCLRCAHAAAVMLDGVQKRYCQQCGKFHVLLDFDEDKRSCRRKLERHNKRRRRK 250
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.7E-26 | 172 | 237 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 28.872 | 173 | 250 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.14E-32 | 175 | 253 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.2E-26 | 176 | 250 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 809 aa Download sequence Send to blast |
MDVSGSGGGG SGSGADAGEH VWDWDNLLDF AFQDDDSLVL PWDDAIGIEA DPTEASPLPT 60 DPTPPQQPVE VEPEPEPVPP SPLQAGGSRR RVRKRDPRLV CPNYLAGRVP CACPEVDEMA 120 AAAEVEDVAT EMLAGARKKS KAAGRGSGAA ARGGGGGGGG GGSGGAGRGA AVEMGCQVSG 180 CEEDIRELKG YHRRHRVCLR CAHAAAVMLD GVQKRYCQQC GKFHVLLDFD EDKRSCRRKL 240 ERHNKRRRRK PDSKGTLEKE IDEQLELSAD GSGDGELREE NIDGTTCEML KTVLSNKVFD 300 RETPVGSEDG LSSPTCIQHN LQNDQNKSIV TFAASVEACL GTKQENAKLT NSPMHDTKSA 360 YSSSCPTGRI SFKLYDWNPA EFPRPLRNQI FEWLSSMPVE LEGYIRPGCT ILTVFIAMPQ 420 HMWDKLSEDA AILVRNLVNA PNSLLLGKGA FFFHVNNMIF QVLKDGATLM STRLEIQAPR 480 IHYVHPTWFE AGKPIELLLC GSSLDQPKFR SLLSFDGNYL KHDCCRLTSH ETFACVENVN 540 PILDSQHEIF QINITQSKPD THGPAFVEVE NMFGLSNFVP ILFGSKQLCF ELERIQDALC 600 GSSYKSVFGE FPGATSDPCD HRELQQTAMS GFLIDIGWLI RKPCPDEFKN VLSLTNIQRW 660 ICVLKFLIQN DFINVLEIIV KSLDNIIGSE ILSNLEMGRL EDHVTAFLGY VRHARNTVNH 720 RAKHDEKTLE TKWGSDSAPN QPNLGTSVPL AKENTDASSE YDLHSTNADC EEEETCVVLF 780 HPHRVGVLAA PVKRYLSSDS TIVDVKLD* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 1e-34 | 176 | 257 | 6 | 87 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 232 | 250 | KRSCRRKLERHNKRRRRKP |
| 2 | 236 | 248 | RRKLERHNKRRRR |
| 3 | 243 | 250 | NKRRRRKP |
| 4 | 244 | 249 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
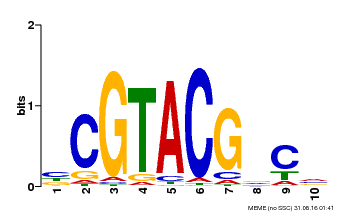
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01001316G0020 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805456.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A3Q8AHQ6 | 0.0 | A0A3Q8AHQ6_9POAL; Squamosa-promoter binding protein-like protein | ||||
| STRING | OGLUM05G17090.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-128 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



