 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01001397G0220 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 290aa MW: 31237 Da PI: 7.6086 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 87.5 | 1.2e-27 | 121 | 180 | 2 | 58 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnh 58
dDg++WrKYGqKe+ g+++pr+YYrCt++ gC ++k+v+r++edp v+++tY+g+H++
PH01001397G0220 121 DDGHSWRKYGQKEILGAKHPRGYYRCTHRnsqGCAATKQVQRTDEDPMVFDVTYHGTHTC 180
8***************************98889*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.3E-28 | 114 | 181 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.009 | 115 | 178 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.01E-25 | 118 | 180 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.7E-39 | 120 | 182 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.9E-27 | 121 | 180 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010193 | Biological Process | response to ozone | ||||
| GO:0042542 | Biological Process | response to hydrogen peroxide | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 290 aa Download sequence Send to blast |
MDCMEGNGGS RLVVTELSHI KELVKQLEVQ LGGSPELCKH LASQIISLTE RSIGMIKSGR 60 FDGRKRSAGD AGLDSPLSAT PSPLSDVSDM PFETKKKRKT MEKRKHQIRV SSAGGAETPI 120 DDGHSWRKYG QKEILGAKHP RGYYRCTHRN SQGCAATKQV QRTDEDPMVF DVTYHGTHTC 180 VQRAMAGQAA AGKSQPVQNP DAHSLLQSLS SSLTVKTEGL AATEPQDWST MPFYFSPVSG 240 LAPEHNPFST PSTSENCFAQ GVSVSPSLVS PATSDSNHLS LAPFPVADAE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 8e-23 | 120 | 180 | 8 | 68 | OsWRKY45 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00525 | DAP | Transfer from AT5G24110 | Download |
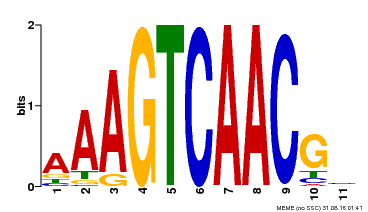
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01001397G0220 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP097435 | 0.0 | FP097435.1 Phyllostachys edulis cDNA clone: bphylf045f19, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020200629.1 | 1e-126 | WRKY transcription factor WRKY62-like | ||||
| TrEMBL | A0A1D8F0I2 | 1e-174 | A0A1D8F0I2_9POAL; WRKY74-1 protein | ||||
| STRING | Traes_5BL_0A3D332A8.1 | 1e-126 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1086 | 38 | 131 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G24110.1 | 2e-36 | WRKY DNA-binding protein 30 | ||||



