 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01001554G0080 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 421aa MW: 45700.9 Da PI: 10.3107 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 94.6 | 1.1e-29 | 26 | 157 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEES.SSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTT.................... CS
HSF_DNA-bind 2 Flkklyeiledeelkelisws.engnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekksks.................... 74
Fl+k+y++++d+++++++sw ++ ++fvv+ + efa+++Lp+yFkh+nf+SFvRQLn+Y + + +
PH01001554G0080 26 FLTKTYQLVDDPATDHIVSWGdDRVSTFVVWRPPEFARDILPNYFKHNNFSSFVRQLNTYVSAAFTCVAGVRVFlefamdgwmdswvltacagg 119
9********************6667***********************************76666666655333678999******99999999 PP
........X..TTSEEEEESXXXXXXXXXXXXXXXXXX CS
HSF_DNA-bind 75 ........k..ekiweFkhksFkkgkkellekikrkks 102
k ++ weF+++ F+kg+k+ll++i+r+k
PH01001554G0080 120 nvvmqgfrKvvPERWEFANEFFRKGEKQLLCEIHRRKM 157
888877661669***********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 2.0E-32 | 15 | 85 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 2.2E-42 | 22 | 155 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 2.81E-25 | 23 | 93 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00447 | 2.1E-26 | 26 | 155 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 3.0E-14 | 26 | 49 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 3.0E-14 | 65 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 3.0E-14 | 78 | 90 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 2.81E-25 | 123 | 155 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 2.0E-32 | 125 | 150 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0008356 | Biological Process | asymmetric cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 421 aa Download sequence Send to blast |
MERCGSWSEC DSAAQAAAQK AVPAPFLTKT YQLVDDPATD HIVSWGDDRV STFVVWRPPE 60 FARDILPNYF KHNNFSSFVR QLNTYVSAAF TCVAGVRVFL EFAMDGWMDS WVLTACAGGN 120 VVMQGFRKVV PERWEFANEF FRKGEKQLLC EIHRRKMTSV SPASPSPSPF FAPPHFALFH 180 PGMAAAQHHQ YVGDDGLVAA HGMGVPFPQP HWREPHAPVA TRLLALAGPV PTPAAEGGGG 240 GRAATAAVLV EENERLRRSN TALLQELAHM RKLYNDIIYF VQNHVRPMAA SPATAAFLQG 300 LGLQARKKPV AANGLNNSGG STTSSSSLTI AEEPSPPPQQ IAADKSGGGR QQQRRSVGAD 360 QALRRAHKRG PVRRGQEEAA FGRRAAAHVA CHQASPCARE RRPKPNHRAV VAAARRLVSS 420 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ldu_A | 2e-15 | 22 | 161 | 17 | 125 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00187 | DAP | Transfer from AT1G46264 | Download |
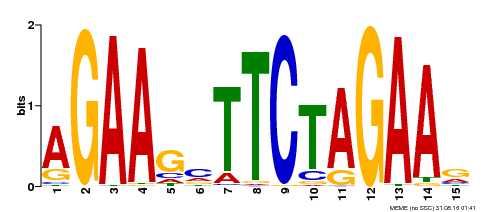
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01001554G0080 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012617 | 0.0 | CP012617.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 9 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015611002.1 | 1e-140 | heat stress transcription factor B-4c | ||||
| Swissprot | Q67U94 | 1e-141 | HFB4C_ORYSJ; Heat stress transcription factor B-4c | ||||
| TrEMBL | A0A0E0M1Z4 | 1e-148 | A0A0E0M1Z4_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC09G11010.1 | 1e-149 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP121 | 37 | 394 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G46264.1 | 5e-69 | heat shock transcription factor B4 | ||||



