 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01001672G0380 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 376aa MW: 41111.2 Da PI: 6.6573 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 35.8 | 1.8e-11 | 320 | 368 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqk 46
r++W++ ++ l+++v+++G+g+Wk I + + R+ ++k+++++
PH01001672G0380 320 RKKWSEMQEKTLIEGVEKYGKGNWKDIKMAYPdvfEDRSTVDLKDKFRN 368
789********************************************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.046 | 315 | 374 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-11 | 316 | 371 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.77E-13 | 317 | 370 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.4E-8 | 319 | 372 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-9 | 320 | 369 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 1.38E-20 | 321 | 371 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 376 aa Download sequence Send to blast |
MPPLAADDAF LILEFVAGNR LVPHPVFTSL LASLPSVSPR TSPRLRKGLA LRALDAALSD 60 ADAHLHKARA VLADPGLASC FPQHPSLSVP DLKRFVDVEW GSLPQSTLEI AADRIAGAGA 120 LDTWANADHA KREKLRLLVG ESTEREILGK LGQYTSAPLP PMAPEINKAA ANAPNTSGAN 180 EAYSAQEDDE ADLVKKNGNV GHAQEDSARH QEDPVKGAPV AQLPENLVTS ATVKGKDHDI 240 PEMINNATAS HVTGQFAPDN NKNHPVPSSK RSLMERNPTA STYEWDGLDD SDPERPSPKR 300 QLPTFEKKQK PSPTFPHKTR KKWSEMQEKT LIEGVEKYGK GNWKDIKMAY PDVFEDRSTV 360 DLKDKFRNME RHQCV* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
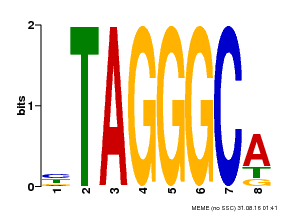
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01001672G0380 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646481.1 | 1e-152 | uncharacterized protein LOC4344404 | ||||
| TrEMBL | A0A446LVA7 | 1e-164 | A0A446LVA7_TRITD; Uncharacterized protein | ||||
| STRING | MLOC_20213.2 | 1e-160 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP13998 | 22 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 9e-11 | TRF-like 5 | ||||



