 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01002143G0030 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1058aa MW: 118284 Da PI: 8.6116 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 163 | 5.4e-51 | 48 | 162 | 4 | 117 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
e +rw +++ei+a+L+n+++ ++++++ ++p+sg+++Ly+rk+vr+frkDG++wkkkkdgktv+E+hekLK+g+ e +++yYa++e++p+f r
PH01002143G0030 48 EaASRWFRPNEIYAVLANHARLKVQAQPINKPSSGTVVLYDRKVVRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEERVHVYYARGEDDPNFFR 141
4489****************************************************************************************** PP
CG-1 97 rcywlLeeelekivlvhylev 117
rcywlL++ele+ivlvhy+++
PH01002143G0030 142 RCYWLLDKELERIVLVHYRQT 162
******************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 72.644 | 42 | 168 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-62 | 45 | 163 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 6.2E-44 | 49 | 161 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.54E-10 | 395 | 483 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 2.60E-16 | 554 | 689 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.8E-17 | 579 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 7.77E-17 | 581 | 695 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.555 | 597 | 701 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.7E-7 | 603 | 693 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.434 | 630 | 662 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 5.4E-6 | 630 | 659 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 280 | 669 | 698 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 6.797 | 778 | 807 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 4.51E-5 | 781 | 847 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 4.4 | 796 | 818 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.529 | 797 | 824 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.059 | 799 | 817 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.08 | 819 | 841 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 820 | 844 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.018 | 822 | 840 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 6.7 | 900 | 922 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.285 | 902 | 930 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.17 | 903 | 922 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1058 aa Download sequence Send to blast |
MASLGARRLA ASPSPGAAAP VRWPRLARRR TPARLSRADL NFEKLKAEAA SRWFRPNEIY 60 AVLANHARLK VQAQPINKPS SGTVVLYDRK VVRNFRKDGH NWKKKKDGKT VQEAHEKLKI 120 GNEERVHVYY ARGEDDPNFF RRCYWLLDKE LERIVLVHYR QTSEENATAP LNAETEVPEV 180 PTINMIQYAS PLTSPDSVSA HTELSCSTAA PEEINSHGGR AISSETDDHG SSLEEINSFW 240 ANLLESSMEN DTSVCGGSLA SSQQIKHDPR DSGNNILNTN TASNAIFVPP LNVTSEAYAT 300 NPGLNQVHAS YFGALKHQED QSQSHLTSDL DSQSEQFVSS LVQTPADGNI PNDVPARQNS 360 LGVWKYLDDD SPCLGDNLTS AIQSFHPVTN ERLFNITEIS PEWAYCSTGN TKVLVVGYFY 420 EQSKHLTESN MYGVFGEKCV AADIVQAGVY RFMAGPHTPG LVNFYLTLDG KTPISEVLSF 480 EYRTIPGSSL MSDLMPLEDE YKKSKLQMQM RLARLLFATN KKKVSPKLLA AGSKVSNLIS 540 ASPEKEWMDL WKTASNSEGM YVSVTEGLLE LVLRNRLQEW LLEKVAEGHK STGRDDLGQG 600 PIHLCSCLGY TWAIRLFGLS GFSLDFRDSS GWTALHWAAC HGRERMVAAL LSAGANPSLV 660 TDPTPESPGG CTAADLAARQ GYDGLAAYLA EKGLTAHFEA MSLFKDTEQS VARTRLTKLQ 720 SEKFENLSEQ ELCLKESLAA YRNAADAASN IQAALRERTL KLQTKAIQLA NPEMEASQIV 780 AAMKIQHAFR NYNRKKEMRA AARIQSHFRT WKIKRNFMNM RRQVIRIQAA YRGHQVRRQY 840 YKVIWSVGVV EKAILRWRKK RRGLRGIGTG MPVAMTVDAE PASTAEEDFF QASRQQAEDR 900 FNRSVVRVQA LFRSYRAQQD YRRMKTAHEE AKSDAARHRS IEPDAVSSPQ RCLGVASISI 960 IETGFNMRLH MRGQRLWEFL TGEFPCTPRP AEPVLPKDAS EDARTKVYES FMDAMETFQT 1020 QYATYKTWID GDARTSAILV TSMEVQFIRD VVGLASA* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
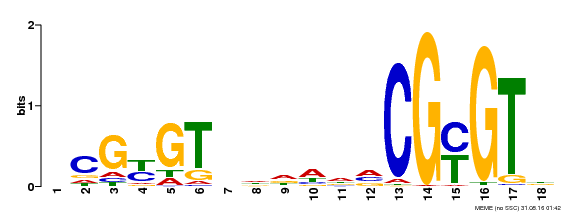
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01002143G0030 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646432.1 | 0.0 | calmodulin-binding transcription activator CBT-like isoform X2 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A0E0I0Z0 | 0.0 | A0A0E0I0Z0_ORYNI; Uncharacterized protein | ||||
| STRING | OS07T0490200-01 | 0.0 | (Oryza sativa) | ||||
| STRING | ONIVA07G13300.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||



