 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01002776G0100 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 303aa MW: 31619.2 Da PI: 6.278 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101 | 6.9e-32 | 121 | 178 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt ++C+vkk++ers+edp+vv++tYeg+H h+
PH01002776G0100 121 EDGYRWRKYGQKAVKNSPYPRSYYRCTNSKCTVKKRLERSSEDPSVVITTYEGQHCHH 178
8******************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 7.3E-33 | 105 | 179 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.57E-27 | 112 | 178 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.505 | 115 | 180 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-36 | 120 | 179 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-24 | 121 | 178 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 303 aa Download sequence Send to blast |
MAGAGVGDWP FSADAYADSS AIFAELGWAA GLDAAGELPP LDPPEATPPP ASEAATPAGS 60 VDGGALSSST DDGATPDDAD GKPAAASTEA ASKPASGKKG QKRARQPRFA FMTKSEIDHL 120 EDGYRWRKYG QKAVKNSPYP RSYYRCTNSK CTVKKRLERS SEDPSVVITT YEGQHCHHTV 180 TFPRGGAGAL AHVHAAALAG QMAFSAHDLY NSLPEMHSTH QNLPPPALDP LVSLVCTPAM 240 SSSSLLQPLH CNQELQATSS YPLSTTTTMS LTSVSTQSSS PASVPVDKGL LDDMVPPAMR 300 HG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 6e-26 | 110 | 177 | 7 | 74 | Probable WRKY transcription factor 4 |
| 2lex_A | 6e-26 | 110 | 177 | 7 | 74 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
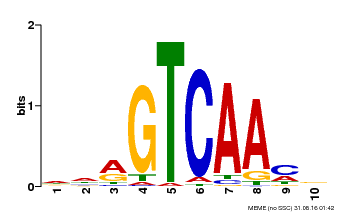
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00067 | PBM | Transfer from AT1G69310 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01002776G0100 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY341859 | 3e-92 | AY341859.1 Oryza sativa (japonica cultivar-group) WRKY19 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020149447.1 | 1e-115 | probable WRKY transcription factor 57 | ||||
| Swissprot | Q9C983 | 4e-55 | WRK57_ARATH; Probable WRKY transcription factor 57 | ||||
| TrEMBL | A0A3B6C5U0 | 1e-119 | A0A3B6C5U0_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A446M9N7 | 1e-119 | A0A446M9N7_TRITD; Uncharacterized protein | ||||
| STRING | Traes_2BL_A69F6C5DF.1 | 1e-120 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2125 | 37 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69310.2 | 2e-53 | WRKY DNA-binding protein 57 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



