 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01002899G0130 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 212aa MW: 23105.4 Da PI: 10.2314 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 103.1 | 1.7e-32 | 108 | 161 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt+ LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
PH01002899G0130 108 PRMRWTTALHARFVHAVELLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 161
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51257 | 5 | 1 | 25 | No hit | No description |
| SuperFamily | SSF46689 | 3.05E-16 | 105 | 161 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-29 | 106 | 161 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.8E-24 | 108 | 161 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.3E-8 | 109 | 160 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 212 aa Download sequence Send to blast |
MLQRNQLPNL SLRISPPAVS SAASCDITTA AKPLPTEPNA ETYAEGSGEV GFFANPSSGA 60 EPPGLSLGLG TPAHADAGRQ SHLQPQGCAF KRAARTSLPG SKRSVRAPRM RWTTALHARF 120 VHAVELLGGH ERATPKSVLE LMNVKDLTLA HVKSHLQMYR TVKGTDRSSS HITEGIPIAF 180 PEEILHCQLP ADRRPCRAKR RWPFYFMPYH E* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-17 | 109 | 163 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-17 | 109 | 163 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-17 | 109 | 163 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-17 | 109 | 163 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-17 | 109 | 163 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates carpel integuments formation. Required for the specification of polarity in the ovule inner integument. Modulates the content of flavonols and proanthocyanidin in seeds. {ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:19054366, ECO:0000269|PubMed:20444210}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00107 | PBM | Transfer from AT5G42630 | Download |
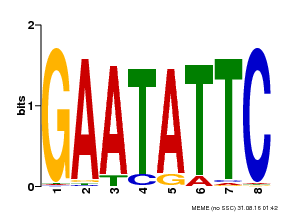
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01002899G0130 |
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP094880 | 0.0 | FP094880.1 Phyllostachys edulis cDNA clone: bphylf040g11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002466395.1 | 3e-78 | probable transcription factor GLK2 | ||||
| Swissprot | Q9FJV5 | 2e-42 | KAN4_ARATH; Probable transcription factor KAN4 | ||||
| TrEMBL | F2ED93 | 2e-81 | F2ED93_HORVV; Predicted protein | ||||
| STRING | Pavir.Ia00330.1.p | 5e-83 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6554 | 37 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42630.2 | 1e-45 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



