 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01002927G0090 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1653aa MW: 184656 Da PI: 8.5376 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.8 | 0.00035 | 1533 | 1556 | 1 | 22 |
EEET..TTTEEESSHHHHHHHHHH CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt 22
++C C++sFs++ +L H r
PH01002927G0090 1533 FSCDieGCDMSFSTQQDLLLHKRD 1556
899999***************985 PP
| |||||||
| 2 | zf-C2H2 | 11.1 | 0.0013 | 1616 | 1642 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y C+ Cg++F+ s++ rH r+ H
PH01002927G0090 1616 YACKepGCGQTFRFVSDFSRHKRKtgH 1642
789999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00558 | 2.0E-5 | 155 | 308 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 15.452 | 155 | 353 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.37E-8 | 170 | 241 | No hit | No description |
| Pfam | PF02373 | 5.4E-13 | 188 | 243 | IPR003347 | JmjC domain |
| Pfam | PF07727 | 2.4E-18 | 484 | 540 | IPR013103 | Reverse transcriptase, RNA-dependent DNA polymerase |
| Pfam | PF07727 | 6.2E-15 | 542 | 611 | IPR013103 | Reverse transcriptase, RNA-dependent DNA polymerase |
| SMART | SM00355 | 9.1 | 1533 | 1555 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.281 | 1556 | 1585 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 10 | 1556 | 1580 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 8.65E-5 | 1557 | 1588 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.5E-4 | 1558 | 1584 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1558 | 1580 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 5.2E-10 | 1585 | 1610 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.427 | 1586 | 1615 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0018 | 1586 | 1610 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1588 | 1610 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.17E-9 | 1596 | 1640 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-9 | 1611 | 1638 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.27 | 1616 | 1642 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.492 | 1616 | 1647 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1618 | 1642 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1653 aa Download sequence Send to blast |
MEIEPEASRY GICKIVPPLP APPREATVQR LKASFTSNAA TSASAGDAVP APTFPTRLQQ 60 VGLSTKNRRA ANRRVWESGE RYTLEAFRSK ARDFELPRHA TPPKNATPLQ LEALFWGACA 120 ARPFNVEYGN DMPGSGFAVP EELELDASNA AAPRDVGETE WNMRVAPRAG GSLLRAMGRD 180 VAGVTTPMLY VAMLYSWFAW HVEDHELHSL NYLHFGKRKT WYGVPRDAML AFEDAVRVHG 240 YADDLNAIKI LRNHDTTRLP QKERLGLAPI LSCLCPISPS PSTAVMNVHL GVASLVNDKV 300 LSCEILLKDA RTFHVTVGVI KLKEEVTHGV STSWYLSKGC KISLKSKIEF ETNGAWLLFQ 360 IPDGFMKSVA FQTLNEKTTV LSPEVLLSAG VPCCRYFSKA DWFKTLENLL SHSLELIILV 420 LATELIFVIR SIVLLAKLIT ISELVQRLLG ILVMIVRAEK RLFLAIAIEA LLVSVLDDTD 480 IFLPSWVYKV KTKSYGSVER YKARLVVKGF QQAYGRDYDK TFAPIAHMTT VCTLIAVAVR 540 GRTLLLLYVD DMLITRDGMD YVAFVKKKLS KKFMMADLGL WSYFMGIEVH SDSDSYYLFQ 600 HRYVQDLLSS FVKSLTSSYL YCSTFSCGSK VLKIGNQYST GHLPSSDYRY NRPDYAAGIT 660 GNYTVTGFNC GEATNIATPR WLQVAKEAAI RRASTNCGPM VSHYQLLYKL ALSLRPREPT 720 NFHTVPRSSR LRDKKKNEGN IMVKEKFVGS VTENNNLLSI LLDKGSCIIV PEIAFPLPSF 780 PMTMEPKVTF KQSLTAGSCS ISQQAAEDMP VDVAADKITG TESMSGSQSA SETSFSACNR 840 RKLYETKYGK VDTAALCLST SEMRSGEIDK DRSHQGGGLL DQGRLPCVQC GILSFACVAI 900 IQPREAAVQF IMSRECISSS AKHGEISKSD DISNWITENR EVVRQQGQAS GTDDNVIHSI 960 SLAQVSDRCR QLYRSDTNGC TSALGLLASA YGSSDSEEET ADNVSTDSEK NDAANQGTNT 1020 RFLGTSVSSS STVDRQKTNS HLYEEECEVR TTASLMKPIE HNSWPITQSS RDTDIGHFTG 1080 LGQPGTSCEQ CPVYLDSVDG LTKSEYPRAE SAAKVIAEEL GMKHDWKDIT FEKATEEDIR 1140 TIQLALQDED AEPTSSDWAV KMGINIYYSA KQSKSPLYSK QIPYNSIIYK AFGQESPDIM 1200 TDYGGQISGT TKKKVAGWWC GKVWMSNQVH PLLAREHEEQ HNHDIVYSKA ILIANSNDKI 1260 QDEPSMRCAT LINRSPSKRI SGRKRGDSID KSRAKKKRCT ACNEATLHCG GLGMNSEARY 1320 HHPGNSDDHD KHEDGDEIEE APSTQQYQQH KLQNMNKKSS SKKQKDDKRN SNFHELYDED 1380 NDVDSWLNLD SDGTTIGNWD NSPQRGLDAV KVKSGGKLQG IKRKSSKGKV SDNLSNGDKK 1440 LQKMNKKSGS KKQKNDKINR QFAEDHNEDN TVNYFHNVER GDEATLDNWD QIPDEKTNDV 1500 KVKSRGKMQS GKRKTSKRQA SEGFRTGDKE PKFSCDIEGC DMSFSTQQDL LLHKRDICPV 1560 KGCNKKFFCH KYLLQHRKVH MDERPLKCAW KGCKKTFKWP WARTEHMRVH TGVRPYACKE 1620 PGCGQTFRFV SDFSRHKRKT GHCCDKIRKS ST* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 8e-57 | 1533 | 1642 | 23 | 132 | Lysine-specific demethylase REF6 |
| 6a58_A | 8e-57 | 1533 | 1642 | 23 | 132 | Lysine-specific demethylase REF6 |
| 6a59_A | 8e-57 | 1533 | 1642 | 23 | 132 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1282 | 1297 | RKRGDSIDKSRAKKKR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
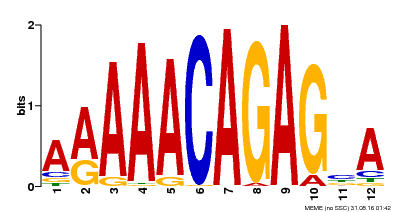
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01002927G0090 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006664493.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6-like | ||||
| TrEMBL | B8BP43 | 0.0 | B8BP43_ORYSI; Uncharacterized protein | ||||
| STRING | OB12G17360.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6718 | 30 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 4e-82 | relative of early flowering 6 | ||||



