 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01005516G0020 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 305aa MW: 33579.6 Da PI: 6.7344 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 46.7 | 5.9e-15 | 198 | 288 | 3 | 90 |
EE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE CS
B3 3 kvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldgrsef 90
k+lt+sdv++ gr+vlpkk ae+ + ++ ++ l++ d +W++k+++ ++ks++y+l + eFvk +gL++gD+++++ ++ +
PH01005516G0020 198 KELTKSDVGNVGRIVLPKKDAESSlpPLFERDPVILQMDDmVLPVTWKFKYRFwpNNKSRMYILD-ATGEFVKTHGLQAGDVFIIYKNS-VPG 288
899******************99999555667777888885555*******99777777777777.9*****************99333.444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.57E-17 | 195 | 284 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 6.4E-22 | 195 | 291 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 4.85E-21 | 195 | 293 | No hit | No description |
| SMART | SM01019 | 1.5E-9 | 196 | 296 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 8.684 | 196 | 298 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.3E-12 | 198 | 289 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 305 aa Download sequence Send to blast |
METEVQRSRS TPDGPGTDAS THNAAQQSPI PRGFGDWSAS SGAFTSLGVQ ATPATSNALH 60 HSLPPCYAFW THYMLNKNAY CSYPPAPHQE HAHTLCHNDN RAKDPGPASS FGIESFTTTS 120 LAPNICAHMP PIEGPISAKE TETSEDLPRV VRSSDERETR NSGELKCETI DTFPESKQGH 180 ESCATKFNSG EYQVILRKEL TKSDVGNVGR IVLPKKDAES SLPPLFERDP VILQMDDMVL 240 PVTWKFKYRF WPNNKSRMYI LDATGEFVKT HGLQAGDVFI IYKNSVPGKF IVRGEKAIQQ 300 TANP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9c_A | 5e-32 | 187 | 284 | 4 | 101 | B3 domain-containing transcription factor LEC2 |
| 6j9c_D | 5e-32 | 187 | 284 | 4 | 101 | B3 domain-containing transcription factor LEC2 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription regulator involved in iron deficiency response and tolerance. May regulate directly iron transporters or other transcription factors involved in iron-deficiency response. Binds specifically to the DNA sequence 5'-CATGC-3' of the IDE1 element found in the promoter of the barley iron deficiency-inducible gene IDS2. {ECO:0000269|PubMed:18025467}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
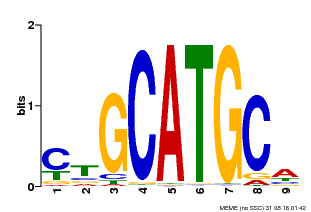
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01005516G0020 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated during the early stage of iron deficiency (at protein level). {ECO:0000269|PubMed:27137867}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020169706.1 | 1e-163 | B3 domain-containing protein IDEF1-like | ||||
| Swissprot | Q6Z1Z3 | 1e-147 | IDEF1_ORYSJ; B3 domain-containing protein IDEF1 | ||||
| TrEMBL | A0A287X0N4 | 1e-167 | A0A287X0N4_HORVV; Uncharacterized protein | ||||
| STRING | EMT27675 | 1e-154 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP11346 | 28 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 2e-35 | B3 family protein | ||||



