 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK04639.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 836aa MW: 96149.6 Da PI: 7.1177 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 88.4 | 1e-27 | 74 | 161 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
+fY+eYAk++GF++++++s++sk+++e+++++f Cs++g + e++ ++r+ ++++t+Cka+++vk+++dgkw ++++ +eHnHel p
PK04639.1 74 SFYQEYAKSMGFTTSIKNSRRSKKSKEFIDAKFACSRYGVTPESDGG---SSRRPSVKKTDCKASMHVKRRSDGKWIIHEFIKEHNHELLP 161
5****************************************999888...7788889*******************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 3.0E-25 | 74 | 161 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 1.8E-25 | 281 | 374 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 10.249 | 561 | 597 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 6.1E-7 | 562 | 596 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.1E-8 | 572 | 599 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 836 aa Download sequence Send to blast |
MDRIQNSVET ALNSRDNANG DRMSEKMIDV VDGMHSRDTG LVSSPKRDLA IFGGDTDFEP 60 CSGIEFESHE AAYSFYQEYA KSMGFTTSIK NSRRSKKSKE FIDAKFACSR YGVTPESDGG 120 SSRRPSVKKT DCKASMHVKR RSDGKWIIHE FIKEHNHELL PALAYHFRIH RNVKLAEKNN 180 IDILHAVSER TRKMYVEMSR KSGGYQNVGS VRAEMNFQFD KGRYLGLDEG DAQVMLEYFK 240 RIQKENPNFF FAIDLNEEQR LRNLFWVDAK SRTDYVSFND VVSFDTSYIK INDKLPFAPF 300 VGVNHHFQPM LLGCAMVADE TKSTFVWLLK TWLRAMHGQA PKVIITDQDK ALKAAIEEVF 360 PDTRHCYSLW QILEKIPETL AHVIKQHENF LPKFNKCIFQ SSTSEQFDMR WWKMVTRFEL 420 QDDEWIRMLY DDRKKWVPTC LGDIFFAGMC TTQRSESMNA FFDKYIHKKI TLKEFVKQYG 480 AILQNRYEEE EIADFDTLHK QPALKSPSPW EKQMSTIYTH AVFKKFQFEV LGVVGCQPKK 540 ESEDETTSTF RVQDCEKDEY FFVTWNETKS DVSCSCHLFE YKGFLCRHAL IVLQMCGLSS 600 IPPQYILKRW TKDAKGRQSR AEGTEQIQTR VQRYNELCKW AIELSEEGSL SEDIYNIAVR 660 ALVEALKNCV NVNNSISSAA ESSGNFLTTC ETEEENQGIL ATKPVRKRNT NRKRKVQAEP 720 DPTLVVAQDS LQQMDNLSSD GITLNGYYGG QQNVQGLLNL MEPPNDSYYV NPPSMQGLGH 780 LNSITPGHDG YYGAQQSIHG LAQLDFRQQT SFSYSLPPDD QRLRSSQLHG SASRPP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
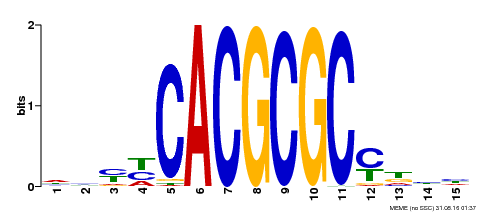
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024019196.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A251Q247 | 0.0 | A0A251Q247_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008229655.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||



