 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK07568.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 837aa MW: 92068.4 Da PI: 6.3161 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.7 | 2e-18 | 16 | 74 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++ +++ps +r++L +++ +++ +q+kvWFqNrR +ek+
PK07568.1 16 KYVRYTPEQVEALERLYHDCPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 74
5679*****************************************************97 PP
| |||||||
| 2 | START | 180.3 | 1.1e-56 | 160 | 368 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEEEEEEXXT CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galqlmvaelq 99
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++++l+v+ ++ g+++l + +l+
PK07568.1 160 IAEETLAEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCTGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRAVDVLNVLPTAngGTIELLYLQLY 258
7899******************************************************.8888888888****************9999*********** PP
TXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHH CS
START 100 alsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaegakt 195
a+++l+p Rdf+++Ry+ l++g++v++++S+ ++q+ p +++vRae+lpSg+li+p+++g+s +++v+h+dl+ ++++++lr+l++s+++ ++kt
PK07568.1 259 APTTLAPaRDFWILRYTSVLEDGSLVVCERSLKNTQNGPAmppVQHFVRAEMLPSGYLIRPCEGGGSIIHIVDHMDLEPWSVPEVLRPLYESSTVLAQKT 358
*************************************99988899******************************************************* PP
HHHHTXXXXX CS
START 196 wvatlqrqce 205
++a+l+++++
PK07568.1 359 TMAALRQLRQ 368
******9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.402 | 11 | 75 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-17 | 13 | 79 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 1.2E-15 | 13 | 79 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.31E-16 | 16 | 76 | No hit | No description |
| Pfam | PF00046 | 5.8E-16 | 17 | 74 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-18 | 18 | 74 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.14E-6 | 68 | 107 | No hit | No description |
| PROSITE profile | PS50848 | 25.437 | 150 | 365 | IPR002913 | START domain |
| CDD | cd08875 | 2.20E-79 | 154 | 370 | No hit | No description |
| SuperFamily | SSF55961 | 1.92E-37 | 159 | 370 | No hit | No description |
| SMART | SM00234 | 1.7E-43 | 159 | 369 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 1.7E-23 | 159 | 366 | IPR023393 | START-like domain |
| Pfam | PF01852 | 4.2E-54 | 160 | 368 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.29E-5 | 395 | 491 | No hit | No description |
| SuperFamily | SSF55961 | 4.29E-5 | 520 | 595 | No hit | No description |
| Pfam | PF08670 | 1.4E-51 | 694 | 836 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 837 aa Download sequence Send to blast |
MAMSCKDGKP GLDNGKYVRY TPEQVEALER LYHDCPKPSS IRRQQLIREC PILSNIEPKQ 60 IKVWFQNRRC REKQRKEASR LQAVNRKLTA MNKLLMEEND RLQKQVSQLV YENGYFRQHT 120 QSTTLATKDT SCESVVTSGQ HHLTPQHPPR DASPAGLLSI AEETLAEFLS KATGTAVEWV 180 QMPGMKPGPD SIGIVAISHG CTGVAARACG LVGLEPTRVA EILKDRPSWF RDCRAVDVLN 240 VLPTANGGTI ELLYLQLYAP TTLAPARDFW ILRYTSVLED GSLVVCERSL KNTQNGPAMP 300 PVQHFVRAEM LPSGYLIRPC EGGGSIIHIV DHMDLEPWSV PEVLRPLYES STVLAQKTTM 360 AALRQLRQIA HEVSQSSVTG WGRRPAALRV LSQRLSRGFN EALNGFTDEG WSMMGNDGMD 420 DVTILVNSSP DKLMGLNLSF ENEFPAVSNA VLCAKASMLL QNVPPAILLR FLREHRSEWA 480 DNNIDAYSAA AVKVGPCSLA GSRVGSFGGQ VILPLAHTIE HDEFLEVIKL EGVSHSPEDA 540 MMPREMFLLQ LCSGMDENAV GSCAELIFAP IDASFADDAP LLPSGFRIIP LDSGKEASSP 600 NRTLDLASAL EIGPNGAKAS SDYSANNGCV RSVMTIAFEF AFESHMQEHV ASMARQYVRS 660 IISSVQRVAL ALSPSHLSSQ AGLRSPLGTP EAQTLARWIC NSYRGYLGVE LLKSSGEGGE 720 TILKTLWHHS DAVMCCSMKA SPVFTFANQA GLDMLETTLV ALQDICLEKI FDDHGRKTLC 780 SEFPQIMQQG FSCLQGGICL SSMGRPVSYE RAVAWKVLNE EENAHCICFM FMNWSFV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
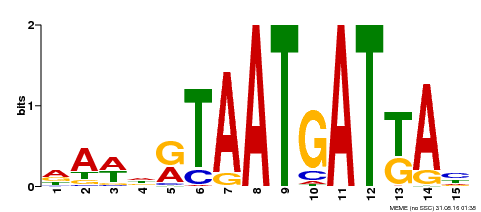
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010098130.1 | 0.0 | homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A2P5ESQ5 | 0.0 | A0A2P5ESQ5_TREOI; Octamer-binding transcription factor | ||||
| STRING | XP_010098130.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1946 | 33 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



