 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK07926.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 591aa MW: 65838.5 Da PI: 5.0339 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.9 | 4.1e-12 | 541 | 589 | 2 | 47 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqky 47
+rW+ E++ l d+vk++G+g+Wk+I +t + Rt+ ++k++w++
PK07926.1 541 KRWSLLEEDALRDGVKKYGKGNWKLILNTYHdlfEERTEVDLKDKWRNM 589
59********************************9************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.646 | 535 | 591 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-11 | 536 | 591 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.74E-14 | 537 | 590 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 0.0015 | 539 | 591 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 4.21E-19 | 541 | 591 | No hit | No description |
| Pfam | PF00249 | 1.8E-8 | 542 | 589 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 591 aa Download sequence Send to blast |
XDMDVSRWIL EFLIRSPGKD RLAKDALSVM PVPNNDLRLN KIALLRTIEG EVSDAMVTET 60 ILEKLELVEA LDSRQESGAA AMADSMKGAY CAVALECTVK FLVSGGGKPG EKYLNALNRV 120 WRGRVRQLEM SGKSKLFTKE LKRCWDEVEA AISDNSVCKK FLVMNTRNEA VRLVLEYLKE 180 AWALLGPSFV EWAAKLTTEH RGLVRLGDGG VRSRGVESVE DEPEVGIERV DEEIEVQEPT 240 AVKLASLGGG VSGMPELISE FRDQEDRTGT SKDQVCNKDE EIEVQEPTAV KLASPGGGLS 300 VNGMPELTSE FRDQEDRTGT SKVCNKDTDV DVTLPDSGTA TFVKEIQSEK VVTRNKQVAI 360 NKRHRGRVRI AEDENFETNE SENRNSISTP EVDRVREALK SSTSELQAVV TDPLPEASRV 420 AEAVATDLAT KNVSHENPLE SQKGTEADAT TNPCIEKNTN SNDPTPGNPS TSHQNNNPQP 480 SLMARNSTAH TYEWDDSIDS IDDSDPRANA SRLNPTRSKR KAVSLLKKDE STKFTKRRKV 540 KRWSLLEEDA LRDGVKKYGK GNWKLILNTY HDLFEERTEV DLKDKWRNMT R |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 518 | 538 | KRKAVSLLKKDESTKFTKRRK |
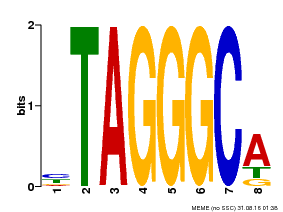
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024029310.1 | 1e-170 | uncharacterized protein LOC21393956 isoform X1 | ||||
| TrEMBL | A0A2P5CYN7 | 0.0 | A0A2P5CYN7_TREOI; GAMYB transcription factor | ||||
| STRING | XP_010108474.1 | 1e-157 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2824 | 32 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 4e-16 | TRF-like 5 | ||||



