 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK08903.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 847aa MW: 97396.8 Da PI: 7.1726 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 85 | 1.2e-26 | 89 | 191 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk.........tekerr.......traetrtgCkaklkvkkekdgkwevtklele 84
+fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ + + +r+ ++t+Cka+++vk+++dgkw+++++++e
PK08903.1 89 SFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYDKSfnrprsrqsK----QdpdnvtgRRSCSKTDCKASMHVKRRSDGKWVIHNFVKE 184
5*******************************************9999999876541....234455569999*************************** PP
FAR1 85 HnHelap 91
HnHel p
PK08903.1 185 HNHELLP 191
****975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 2.1E-24 | 89 | 191 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 1.2E-28 | 289 | 381 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.619 | 569 | 605 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 9.2E-6 | 577 | 604 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.1E-8 | 580 | 607 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 847 aa Download sequence Send to blast |
MDIDLRLPSG EHDKEGEEST AIDNMLDGEE KLHNGDIISG NMVDIGDEVH TEDGGDLNSP 60 TANMVVFKED TNLEPLSGME FESHGEAYSF YQEYARSMGF NTAIQNSRRS KTSREFIDAK 120 FACSRYGTKR EYDKSFNRPR SRQSKQDPDN VTGRRSCSKT DCKASMHVKR RSDGKWVIHN 180 FVKEHNHELL PAQAVSEQTR KMYAAMARQF AEYKSVVGLK NDPKNPFEKG RNLAVEAGDL 240 KILLDFFTQM QNVNANFFYA VDVGEDQRIK NLFWVDAKSR HDYANFSDVV SFDTTYIRNK 300 YKMPLALFIG VNQHYQFMLL GCALISDESV TTYSWLMQTW LKAMGGQVPK VIITDHDKVL 360 KSVISEVFPI VHHCFFLWHI LGRVSETLGH VIKQHENFMA KFEKCIYRSW TIEKFDKRWW 420 KILDKFELRG DEWIHSLYED RKQWVPSFMR DALLAGMSTV QRSDSVNYYF DKYVHKKTSV 480 QEFLKQHETI LQDRYEEEAK ADSDTWNKQP TLKSPSPLEK SVSGVYTHAV FKKFQVEVLG 540 AVACIPKKER QDEISIIFQV QDFEKGQDYQ VSWSEMKSEV SCLCRLFEYK GYLCRHALIV 600 LQMCGLSVIP PQYILKRWTR DAKNRHLTGE ESGQLQTRVQ RYNDLCQRAM KLCEEGSLSQ 660 ESYSIACRAL EETFGNCTSV NNSSRSLVDA GTSATHGILC MEEDSQSKNI VKTNKKKNPT 720 KKRKVSFEPD VITVGTQDSL QQMDNKLNPR AVTLDSYYGA QQNVQGMVQL NLMAPRDNYY 780 GNQQTIQGLG QLNSIAPSHD GYYSAQQSMH ALGQMEFFSR PSFTYGIRQD DPNVRTTSLH 840 DDASRHG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
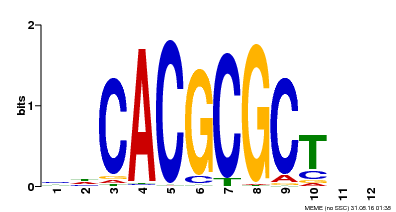
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024019194.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A251Q297 | 0.0 | A0A251Q297_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008229656.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



