 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK10342.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 148aa MW: 16562.2 Da PI: 9.5011 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 109.9 | 1.3e-34 | 83 | 137 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+rlrWt+eLHerFv+av+qLGG+++AtPk +l++m+v+gLt++hvkSHLQkYRl
PK10342.1 83 KQRLRWTHELHERFVDAVAQLGGPDRATPKGVLRVMGVQGLTIYHVKSHLQKYRL 137
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.485 | 80 | 140 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.79E-18 | 82 | 138 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-33 | 82 | 138 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.9E-26 | 83 | 138 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.2E-10 | 85 | 136 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 148 aa Download sequence Send to blast |
XYIHNNHTHT YSFIITHEKT QASIVPNRRA GAWTETISVR FQICEAAPST DICVHGRHLD 60 SGVNTMDPVN GGNSLNNPSL ASKQRLRWTH ELHERFVDAV AQLGGPDRAT PKGVLRVMGV 120 QGLTIYHVKS HLQKYRLAKY LPESSSDX |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 9e-26 | 83 | 140 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 9e-26 | 83 | 140 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 9e-26 | 83 | 140 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 9e-26 | 83 | 140 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00628 | PBM | 25215497 | Download |
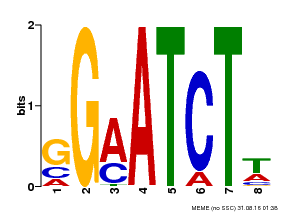
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010096513.1 | 1e-58 | myb family transcription factor PHL7 isoform X1 | ||||
| Refseq | XP_021637722.1 | 4e-59 | myb family transcription factor PHL7-like | ||||
| Refseq | XP_024021465.1 | 1e-58 | myb family transcription factor PHL7 isoform X2 | ||||
| Swissprot | Q9SJW0 | 4e-42 | PHL7_ARATH; Myb family transcription factor PHL7 | ||||
| TrEMBL | W9RQE9 | 3e-57 | W9RQE9_9ROSA; Myb family transcription factor APL | ||||
| STRING | XP_010096513.1 | 6e-58 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8610 | 33 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01060.1 | 2e-44 | G2-like family protein | ||||



