 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK12461.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 267aa MW: 30170.9 Da PI: 9.7988 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 167.3 | 5.1e-52 | 14 | 138 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk.kgel 99
lppGfrFhPtdeelvv+yLk+kv + +l++ ++i+evd++k++PwdLp e+e yfFs+r+ ky++g+r+nrat sgyWkatg dk++l++ ++++
PK12461.1 14 LPPGFRFHPTDEELVVQYLKRKVFSYPLPA-SIIPEVDVCKSDPWDLPGD---LEQERYFFSTREAKYPNGNRSNRATGSGYWKATGIDKQILTSrGNHV 109
79****************************.89***************54...46799**********************************99857788 PP
NAM 100 vglkktLvfykgrapkgektdWvmheyrl 128
vg+kktLvfy+g+ p+g++tdW+mheyrl
PK12461.1 110 VGMKKTLVFYRGKPPQGTRTDWIMHEYRL 138
***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 6.02E-60 | 10 | 176 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.946 | 14 | 176 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.0E-27 | 15 | 138 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 267 aa Download sequence Send to blast |
MEKLNFVKNG VLRLPPGFRF HPTDEELVVQ YLKRKVFSYP LPASIIPEVD VCKSDPWDLP 60 GDLEQERYFF STREAKYPNG NRSNRATGSG YWKATGIDKQ ILTSRGNHVV GMKKTLVFYR 120 GKPPQGTRTD WIMHEYRLAN TTNNPQTKAT PEKSLHQSPV VVAPMESWVL CRIFLKKRGN 180 KNEDENLQTG VDHIVVRKVK STTKPVFYDF MTKEKKRADL NLAPTSSSSG SSGVTQVSSN 240 ESEEHEESSS CNSFPYFXKK AAAAGVE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 6e-52 | 12 | 177 | 15 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 6e-52 | 12 | 177 | 15 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 6e-52 | 12 | 177 | 15 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 6e-52 | 12 | 177 | 15 | 166 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 6e-52 | 12 | 177 | 18 | 169 | NAC domain-containing protein 19 |
| 4dul_A | 6e-52 | 12 | 177 | 15 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 6e-52 | 12 | 177 | 15 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that negatively regulates the expression of genes involved in xylem vessel formation. Represses the transcriptional activation activity of NAC030/VND7, which regulates protoxylem vessel differentiation by promoting immature xylem vessel-specific genes expression (PubMed:20388856). Transcriptional activator that regulates the COLD-REGULATED (COR15A and COR15B) and RESPONSIVE TO DEHYDRATION (LTI78/RD29A and LTI65/RD29B) genes by binding directly to their promoters. Mediates signaling crosstalk between salt stress response and leaf aging process (PubMed:21673078). May play a role in DNA replication of mungbean yellow mosaic virus (PubMed:24442717). {ECO:0000269|PubMed:20388856, ECO:0000269|PubMed:21673078, ECO:0000269|PubMed:24442717}. | |||||
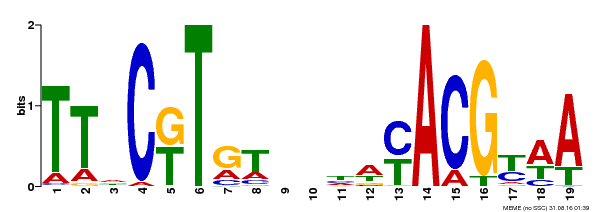
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00507 | DAP | Transfer from AT5G13180 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA) and salt stress. {ECO:0000269|PubMed:21673078}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015881782.1 | 1e-147 | NAC domain-containing protein 83-like | ||||
| Refseq | XP_015902286.1 | 1e-147 | NAC domain-containing protein 83-like | ||||
| Swissprot | Q9FY93 | 1e-107 | NAC83_ARATH; NAC domain-containing protein 83 | ||||
| TrEMBL | A0A2P5BSW6 | 1e-154 | A0A2P5BSW6_PARAD; NAC domain containing protein | ||||
| STRING | cassava4.1_014491m | 1e-141 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF704 | 34 | 139 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13180.1 | 1e-103 | NAC domain containing protein 83 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



