 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK13281.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 140aa MW: 16767.2 Da PI: 10.7668 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.2 | 9.2e-19 | 18 | 65 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd ll++++ ++G g+W++ a++ g++Rt+k+c++rw++yl
PK13281.1 18 RGPWTLEEDTLLIHYISLHGEGHWNLLAKRAGLKRTGKSCRLRWLNYL 65
89*********************************************7 PP
| |||||||
| 2 | Myb_DNA-binding | 53.6 | 5e-17 | 71 | 114 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ T++E++l+++++ ++G++ W++Ia++++ gRt++++k++w++
PK13281.1 71 RGNLTPQEQLLILELHSKWGNR-WSRIAQHLP-GRTDNEIKNYWRT 114
7999******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.348 | 13 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.95E-31 | 16 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.0E-16 | 17 | 67 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-17 | 18 | 65 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.4E-24 | 19 | 72 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.47E-10 | 20 | 65 | No hit | No description |
| PROSITE profile | PS51294 | 25.445 | 66 | 120 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.7E-15 | 70 | 118 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.3E-16 | 71 | 114 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.7E-24 | 73 | 119 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.19E-10 | 75 | 114 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 140 aa Download sequence Send to blast |
MHTITTKFNN EEEIELRRGP WTLEEDTLLI HYISLHGEGH WNLLAKRAGL KRTGKSCRLR 60 WLNYLKPDIK RGNLTPQEQL LILELHSKWG NRWSRIAQHL PGRTDNEIKN YWRTRVQKQA 120 RQLNIESNSK KFIEAVRGFW |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-26 | 15 | 120 | 4 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor of phosphate (Pi) starvation-induced genes. Regulates negatively Pi starvation responses via the repression of gibberellic acid (GA) biosynthesis and signaling. Modulates root architecture, phosphatase activity, and Pi uptake and accumulation. {ECO:0000269|PubMed:19529828}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00159 | DAP | Transfer from AT1G25340 | Download |
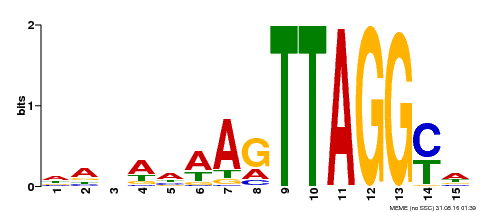
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid (PubMed:16463103). Induced reversibly in response to phosphate (Pi) deficiency but repressed in the presence of Pi, specifically in the leaves. Availability of Pi increases with decreased levels (PubMed:19529828). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:19529828}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010099251.1 | 2e-87 | transcription factor MYB62 | ||||
| Swissprot | Q9C9G7 | 1e-76 | MYB62_ARATH; Transcription factor MYB62 | ||||
| TrEMBL | A0A2P5F0S4 | 2e-88 | A0A2P5F0S4_TREOI; MYB transcription factor | ||||
| TrEMBL | J9JF55 | 2e-88 | J9JF55_HUMLU; Myb transcription factor | ||||
| STRING | XP_010099251.1 | 7e-87 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1639 | 34 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25340.1 | 3e-80 | myb domain protein 116 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



