 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK13838.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 173aa MW: 19227.9 Da PI: 9.7519 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 151.7 | 6.9e-47 | 3 | 154 | 3 | 163 |
YABBY 3 vfssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkeelleelkveeenlksnvekeesastsvssekls 102
+ s+++Cyv+CnfC+t+lav++P++ + +vtv+CGhC++l + +++ ++ + h ++ ++ + + ++ + ++ s+s s +s++ s
PK13838.1 3 FTPPSDHLCYVRCNFCTTVLAVALPYKRFLDTVTVKCGHCSNLSFL--STR--PTPLQPH---QFLDQ-YPLMTHLPGFCNNDHLRKGSSSGSSPSSTTS 94
56789************************************98653..333..3444444...33332.1222223333444455555555555655544 PP
YABBY 103 enedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahf 163
+ ++ +p v++PPek+ r Psaynrf+keeiqrik++nP+i hreafsaaaknWa +
PK13838.1 95 GDPMSST-GAPFVVKPPEKKHRLPSAYNRFMKEEIQRIKSANPEIPHREAFSAAAKNWARY 154
4444444.46789**********************************************75 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 1.9E-52 | 6 | 154 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 2.09E-8 | 100 | 152 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 3.8E-5 | 108 | 153 | IPR009071 | High mobility group box domain |
| CDD | cd01390 | 6.35E-6 | 115 | 151 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010254 | Biological Process | nectary development | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0048479 | Biological Process | style development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 173 aa Download sequence Send to blast |
MEFTPPSDHL CYVRCNFCTT VLAVALPYKR FLDTVTVKCG HCSNLSFLST RPTPLQPHQF 60 LDQYPLMTHL PGFCNNDHLR KGSSSGSSPS STTSGDPMSS TGAPFVVKPP EKKHRLPSAY 120 NRFMKEEIQR IKSANPEIPH REAFSAAAKN WARYIPHPPA GSVSGSNKRI LQQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor required for the initiation of nectary development. Also involved in suppressing early radial growth of the gynoecium, in promoting its later elongation and in fusion of its carpels by regulating both cell division and expansion. Establishes the polar differentiation in the carpels by specifying abaxial cell fate in the ovary wall. Regulates both cell division and expansion. {ECO:0000269|PubMed:10225997, ECO:0000269|PubMed:10225998, ECO:0000269|PubMed:10535738, ECO:0000269|PubMed:11714690, ECO:0000269|PubMed:15598802, ECO:0000269|Ref.10}. | |||||
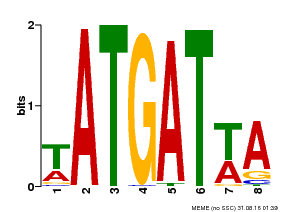
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00220 | DAP | Transfer from AT1G69180 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by SPT and by A class genes AP2 and LUG in the outer whorl. In the third whorl, B class genes AP3 and PI, and the C class gene AG act redundantly with each other and in combination with SEP1, SEP2, SEP3, SHP1 and SHP2 to activate CRC in nectaries and carpels. LFY enhances its expression. {ECO:0000269|PubMed:10225998, ECO:0000269|PubMed:15598802}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024018325.1 | 1e-79 | protein CRABS CLAW | ||||
| Swissprot | Q8L925 | 3e-57 | CRC_ARATH; Protein CRABS CLAW | ||||
| TrEMBL | A0A2P5E2G7 | 3e-83 | A0A2P5E2G7_PARAD; YABBY protein | ||||
| STRING | XP_004509753.1 | 4e-72 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF10259 | 30 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69180.1 | 2e-58 | YABBY family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



