 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK16724.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 243aa MW: 27074.9 Da PI: 10.0966 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 122.4 | 5.2e-38 | 22 | 195 | 2 | 134 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt.................................... 65
g+kdrhsk+ T++g+RdRRvRls+++a+ f+dLqd+LGfd++sk++eWL+++a +ai el+++
PK16724.1 22 SGGKDRHSKVWTSKGLRDRRVRLSVTTAIHFYDLQDRLGFDQPSKAVEWLINAASEAIAELPSLngsfpdtpkqlsdekrgsvgtntgggeqgfdsaeve 121
689******************************************************************9999999999999999999999855544443 PP
TCP 66 ....ssssasec...eaesssssasnsssgkaaksaakskksqksaasalnlak....esrakarararertrekmrikn 134
+ ++ + ++++n+++++ +++a+s+ s++s+ s l+l++ r kar+rarer ++++ ++n
PK16724.1 122 mdgiN------YhhhQHLQLDQQNQNQNQNQPLSKSACSSTSETSKGSGLSLSRsdirVNRVKARERARERAAKEKEKEN 195
33221......022345555555556666666777777777777777778887754446669********7777666654 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 1.8E-44 | 23 | 193 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 30.842 | 24 | 82 | IPR017887 | Transcription factor TCP subgroup |
| PROSITE profile | PS51370 | 9.946 | 173 | 191 | IPR017888 | CYC/TB1, R domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:1903508 | Biological Process | positive regulation of nucleic acid-templated transcription | ||||
| GO:2000306 | Biological Process | positive regulation of photomorphogenesis | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
XGANRLRGWH HSSRIIRVSR ASGGKDRHSK VWTSKGLRDR RVRLSVTTAI HFYDLQDRLG 60 FDQPSKAVEW LINAASEAIA ELPSLNGSFP DTPKQLSDEK RGSVGTNTGG GEQGFDSAEV 120 EMDGINYHHH QHLQLDQQNQ NQNQNQPLSK SACSSTSETS KGSGLSLSRS DIRVNRVKAR 180 ERARERAAKE KEKENESRIV VHHNDNNNNN NNNLNSISQN STFTELLTAG MSNNSNNNNN 240 NNX |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 3e-23 | 29 | 83 | 1 | 55 | Putative transcription factor PCF6 |
| 5zkt_B | 3e-23 | 29 | 83 | 1 | 55 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Plays a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164). Participates in ovule develpment (PubMed:25378179). Participates in ovule develpment (PubMed:25378179). Promotes light-regulated transcription of CHS, CAB, HYH and HY5. Regulates positively photomorphogenesis (e.g. hypocotyl elongation inhibition and cotyledon opening in response to blue light) (PubMed:26596765). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:25378179, ECO:0000269|PubMed:26596765}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00063 | PBM | Transfer from AT4G18390 | Download |
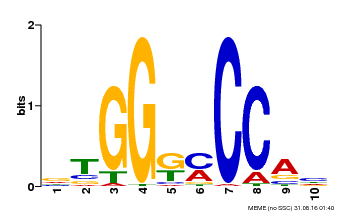
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW (PubMed:12931144). Induced by blue light. Stabilized by light but labile in darkness due to proteasome-dependent proteolysis (at protein level) (PubMed:26596765). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:26596765}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001306852.1 | 1e-106 | transcription factor TCP2-like | ||||
| Refseq | XP_012072140.1 | 1e-106 | transcription factor TCP2-like isoform X1 | ||||
| Refseq | XP_012072141.1 | 1e-106 | transcription factor TCP2-like isoform X1 | ||||
| Refseq | XP_012072142.1 | 1e-106 | transcription factor TCP2-like isoform X1 | ||||
| Refseq | XP_012072143.1 | 1e-106 | transcription factor TCP2-like isoform X1 | ||||
| Refseq | XP_020534893.1 | 1e-106 | transcription factor TCP2-like isoform X1 | ||||
| Swissprot | Q93V43 | 4e-53 | TCP2_ARATH; Transcription factor TCP2 | ||||
| TrEMBL | A0A067L0H7 | 1e-104 | A0A067L0H7_JATCU; TCP transcription factor BRC1 | ||||
| STRING | POPTR_0004s06440.1 | 1e-103 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4019 | 31 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18390.2 | 4e-43 | TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



