 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK18992.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 351aa MW: 39635.3 Da PI: 4.9867 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.3 | 6.3e-19 | 57 | 110 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ +q+++Le+ Fe ++++ e++ +LA++lgL+ rqV vWFqNrRa++k
PK18992.1 57 KKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWK 110
566899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.9 | 2.1e-41 | 56 | 148 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrls +qvk+LE++Fe e+kLeperKv+la+eLglqprqvavWFqnrRAR+ktkqlE+dy +Lk++ydalk + ++L++++e+L +e++e
PK18992.1 56 EKKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLERDYGHLKANYDALKLSYDTLKHDNESLVKEIRE 148
69**************************************************************************************99986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.69E-19 | 45 | 114 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.443 | 52 | 112 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.3E-18 | 55 | 116 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.01E-16 | 57 | 113 | No hit | No description |
| Pfam | PF00046 | 4.1E-16 | 57 | 110 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 9.3E-20 | 59 | 119 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.1E-5 | 83 | 92 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 87 | 110 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.1E-5 | 92 | 108 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.0E-17 | 112 | 154 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 351 aa Download sequence Send to blast |
MKRLSSDDSL GALMSVCPTT EEHSPSSNHI YGREFRSMLD GLDEEGCVEE SGHVSEKKRR 60 LSVDQVKALE KNFEVENKLE PERKVKLAQE LGLQPRQVAV WFQNRRARWK TKQLERDYGH 120 LKANYDALKL SYDTLKHDNE SLVKEIRELK SKLQDEKSES NNLSVKEELV LIHNIGPTES 180 ENKQAVPHPD HHHHHNNHVL PSEECKELNF ESLNDTNNGV VLEVSSMFPD FNKDGSSDSD 240 SSAILNDDIN LNSNNNNSSC SPNAAISSTA LLQTPAASAS ASLNFNCSSS SNNNNNNNNN 300 NSSNCFQFQK AASFQPQFVK IEEHNFFAEE ACNFFSDDQA PTLQWYCSDQ W |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 104 | 112 | RRARWKTKQ |
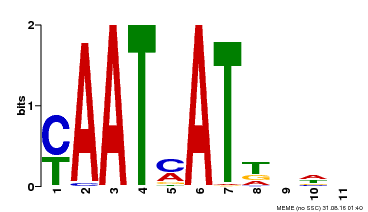
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235215 | 3e-40 | AC235215.1 Glycine max strain Williams 82 clone GM_WBb0020M12, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016651837.1 | 1e-146 | PREDICTED: homeobox-leucine zipper protein ATHB-6-like isoform X2 | ||||
| TrEMBL | A0A2P5AY58 | 1e-170 | A0A2P5AY58_PARAD; Homeobox protein | ||||
| STRING | XP_008235150.1 | 1e-144 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1868 | 34 | 89 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 3e-66 | homeobox protein 6 | ||||



