 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PK24580.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Cannabis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 316aa MW: 34700.4 Da PI: 4.4644 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.9 | 2.8e-19 | 105 | 154 | 1 | 54 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkle 54
s y+GVr++k +g+W+AeIrd + + r +lg++ t+eeA ka++a r+++e
PK24580.1 105 SPYRGVRQRK-WGKWAAEIRDpF-----KgSRLWLGTYTTPEEASKAYEAKRLEFE 154
68*******9.**********55.....45***********************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.83E-16 | 105 | 153 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 19.954 | 106 | 163 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 9.5E-19 | 106 | 168 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.27E-22 | 107 | 153 | No hit | No description |
| PRINTS | PR00367 | 1.3E-9 | 107 | 118 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.9E-12 | 107 | 155 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.0E-22 | 107 | 154 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-9 | 129 | 145 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 316 aa Download sequence Send to blast |
MPEPRNQSSK SMRKIRVICS DPYATDSSSS EDEGFGRKIS KKCKRIVKEI HVPLFPTKPT 60 KTLESESSVQ DSNYGGCVVV SQTLSNPKRR VFAKTSNGSR KPSSSPYRGV RQRKWGKWAA 120 EIRDPFKGSR LWLGTYTTPE EASKAYEAKR LEFEAMANAN STPKFNSSSS SASASVSASV 180 SVSVSVSASA EDSESVLSHN SPASVLELET SASDSKLDET INANVTEEET PDLGFMDEVL 240 ASFPPLEQDI NFGPELDTLF LDDIDDLLNG FNNIDDIQLF GVEDKDSSNL PNYDFDDLGK 300 DDISFWMEGA LNIPCL |
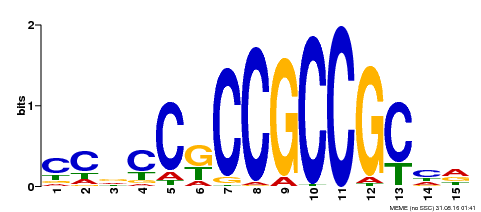
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00217 | DAP | Transfer from AT1G68550 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023885153.1 | 2e-98 | ethylene-responsive transcription factor ERF119-like | ||||
| TrEMBL | A0A2P5DZ65 | 1e-136 | A0A2P5DZ65_PARAD; AP2/ERF transcription factor | ||||
| STRING | XP_010103775.1 | 2e-93 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2690 | 34 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68550.2 | 7e-34 | ERF family protein | ||||



