 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00003641-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 466aa MW: 50774.3 Da PI: 4.8709 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.8 | 1.6e-19 | 108 | 157 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+G+r+++ +g+W+AeIrdp++ r++lg+f taeeAa+a++a +kk++g
PSME_00003641-RA 108 VYRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTFNTAEEAARAYDAEAKKIRG 157
69*******.**********954...4*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 23.657 | 108 | 165 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.2E-31 | 108 | 166 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.4E-12 | 108 | 157 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.73E-22 | 108 | 166 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 6.8E-35 | 108 | 171 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.6E-11 | 109 | 120 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.62E-33 | 109 | 167 | No hit | No description |
| PRINTS | PR00367 | 1.6E-11 | 131 | 147 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 466 aa Download sequence Send to blast |
MCGGAIISDF IPASRSRRVS ARDLWPDFDT FAEFINGGAV PESFNKSASF DDGVPEFDGE 60 YKQKNKPRKD FAKSKQGFSS GSESETIPPL KDFKVPAAKS TGRKRKNVYR GIRQRPWGKW 120 AAEIRDPRKG VRVWLGTFNT AEEAARAYDA EAKKIRGKKA KLNFADDSFA AKADISMKVS 180 GKKVKSCAKN IDLLLEGLNV NSKVKPSYSA NPDLLEGYNI NREVKLSLKD VCRSDLPIYG 240 HNDMEYGDFE FLKPAASVQG NSNVSAGQSS ENSNLSQTLQ KSCSCEICSH NYSEASNLMP 300 TAYGDAVNFE QVNSSNPAGY FDSDHSSMSV DGAHIPWAHE TKTPEITSVY DDANESTFVD 360 IKPPVVGAIK DGSVDTPIEV DGGTGECYKT KDDIVQLEFS EELSALESYL GLAVGSNLKA 420 TADQSMDDAG QVEGSFSDEQ RSLEQWIYGD LSISGLAFPS SLGSPF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 1e-20 | 109 | 164 | 3 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
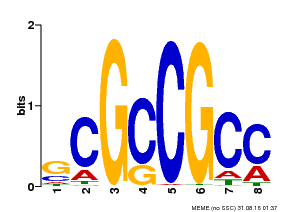
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G53910.2 | 1e-31 | related to AP2 12 | ||||



