 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00004812-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 472aa MW: 52347.4 Da PI: 4.677 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.3 | 1.2e-20 | 175 | 225 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGVr+++ +g+W+AeIrdps+ r +lg++ taeeAa+a+++a+ k++g
PSME_00004812-RA 175 SKYKGVRQRR-WGKWAAEIRDPSK---GVRLWLGTYNTAEEAAQAYDKAAMKING 225
89*****998.**********965...4***********************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 4.9E-22 | 175 | 233 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 7.7E-14 | 175 | 225 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.93E-33 | 175 | 232 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 3.1E-30 | 176 | 233 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.6E-36 | 176 | 239 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.551 | 176 | 233 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.1E-9 | 177 | 188 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.1E-9 | 199 | 215 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 472 aa Download sequence Send to blast |
MPGPQIFPQS LCTTALKTSK KVRGQNKLNT QYVIENERVE RSLATVTRTV RVICTDPDAT 60 DSSSDEDEEA VRFEPKRVVR EIHIPADCSP SVSDSEDEFQ LPICSSLSSL CRIEEECKDV 120 CQEFYAPPEK KLRKNTKGKK VSRKQRTVLK AFERVKSGRV SKCSSSLRSF ADKTSKYKGV 180 RQRRWGKWAA EIRDPSKGVR LWLGTYNTAE EAAQAYDKAA MKINGPSATT NFSGSCTSYT 240 SHRNPPRAAK KQIYDRQWIA AHSVMTRSSA NAKMKPITVE SHMTSSSASL SCISEEGSEE 300 GSDEDWIPVS SLASAPTVST SYSLIDDVGS EDASCSLMME HSTSCSISSD LLADCLEDNA 360 YHGSKNECLA NLDNCNFLLD GTLDLIEHLS IENEDNPLVD FLIPPIHEQN VLESTYPDSY 420 FLDEFGQVFD IGDAGPKDIV SLPDDLDLMD EQNAIVSLNF DLDAEAMSWM NL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 2e-19 | 173 | 232 | 2 | 62 | ATERF1 |
| 3gcc_A | 2e-19 | 173 | 232 | 2 | 62 | ATERF1 |
| Search in ModeBase | ||||||
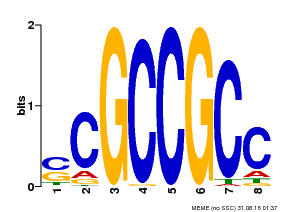
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT118738 | 0.0 | BT118738.1 Picea glauca clone GQ04009_L20 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0C9RXU0 | 1e-150 | A0A0C9RXU0_9SPER; TSA: Wollemia nobilis Ref_Wollemi_Transcript_5206_2819 transcribed RNA sequence | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 2e-18 | cytokinin response factor 2 | ||||



