 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00009599-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 755aa MW: 82609.1 Da PI: 6.0319 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 67.2 | 2.1e-21 | 111 | 166 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r +++k+l+L++rqVk+WFqNrR+++k
PSME_00009599-RA 111 KKRYHRHTPQQIQELEALFKECPHPDEKQRLDISKRLNLETRQVKFWFQNRRTQMK 166
688999***********************************************999 PP
| |||||||
| 2 | START | 223.1 | 8.6e-70 | 295 | 518 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kae 80
ela++ ++el+k+a+a+e +W s e +n++e++++f+++ + +ea r++g+v+ ++ +lve+l+d+ +W+e+++ +a+
PSME_00009599-RA 295 ELALASMDELFKMAQADETLWIPSLdagkETLNYEEYMRQFPSNITpkpigLATEATRETGMVITNSLNLVETLMDVD-RWKEMFPcmisRAA 386
57899********************9***************99999********************************.************** PP
EEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEE CS
START 81 tlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtw 166
+++vissg galqlm+aelq+lsplvp R+++f+R+++q+ +g+w++vdvS+d+ ++++ s+++++++lpSg+li++++ng+skvtw
PSME_00009599-RA 387 MIDVISSGmggtrnGALQLMYAELQVLSPLVPsREVYFLRFCKQHAEGVWAVVDVSIDNLRENS-PSGFMKCRRLPSGCLIQDMPNGYSKVTW 478
****************************************************************.7*************************** PP
EE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 167 vehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
veh+++++r +h l+rsl++sg+a+ga++w+atlqrqce+
PSME_00009599-RA 479 VEHAEYDDRGVHRLYRSLLNSGMAFGAQRWLATLQRQCEC 518
**************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.92E-20 | 93 | 168 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-21 | 96 | 162 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.313 | 108 | 168 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.2E-19 | 109 | 172 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.28E-19 | 110 | 168 | No hit | No description |
| Pfam | PF00046 | 5.8E-19 | 111 | 166 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 143 | 166 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 46.607 | 286 | 521 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.61E-41 | 289 | 518 | No hit | No description |
| CDD | cd08875 | 6.38E-131 | 290 | 517 | No hit | No description |
| Pfam | PF01852 | 4.4E-60 | 295 | 518 | IPR002913 | START domain |
| SMART | SM00234 | 1.4E-60 | 295 | 518 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 1.2E-5 | 380 | 513 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 2.93E-17 | 547 | 747 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 755 aa Download sequence Send to blast |
MSFGGFLDSG SGGKLISASD HNMASGAIAQ PRLVSPGVMP MARATKNIYT SPGLSLALTS 60 GIEGQGEVGA SVGECEQGKS KDEEYESRSG SDNMDGGSGD EQDAADNPPR KKRYHRHTPQ 120 QIQELEALFK ECPHPDEKQR LDISKRLNLE TRQVKFWFQN RRTQMKTQLE RHENSILRQE 180 NEKLRSENLS IRDAMRNPIC TNCGGPAVLG EMSFEEQQLR IENARLKEEL DRLCALAGKF 240 FGRPIHSMPS VPHIPKSSLD LGVGGMPSAG ADLMHGHAGG RTGNMIGIER SMLAELALAS 300 MDELFKMAQA DETLWIPSLD AGKETLNYEE YMRQFPSNIT PKPIGLATEA TRETGMVITN 360 SLNLVETLMD VDRWKEMFPC MISRAAMIDV ISSGMGGTRN GALQLMYAEL QVLSPLVPSR 420 EVYFLRFCKQ HAEGVWAVVD VSIDNLRENS PSGFMKCRRL PSGCLIQDMP NGYSKVTWVE 480 HAEYDDRGVH RLYRSLLNSG MAFGAQRWLA TLQRQCECLA ILIATANVPR DPTAIPTPNG 540 RRSMLRLAQR MTDNFCAGVS ASTVHTWNKL SGNIDDDVRV MTRKSVDDPG EPPGVVLSAA 600 TSVWLPVSPQ RLFDFLRDER LRSEAMNSNQ SSSMLILQET CTDASGSLVV YAPVDIPAMH 660 VVMSGGDPAY VALLPSGFAI LPDGPECRPL ALNPTGNGVG VNSPRVGGSL LTVAFQILVN 720 SLPTAKLTVE SVETVNNLIS CTVQKIKAAL HCEDA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
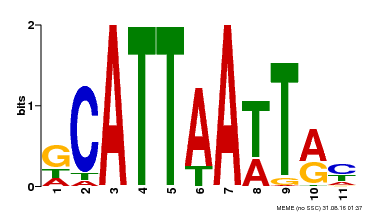
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF328842 | 0.0 | AF328842.1 Picea abies homeodomain protein HB2 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002272264.2 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Refseq | XP_010661561.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | Q8S555 | 0.0 | Q8S555_PICAB; Homeodomain protein HB2 | ||||
| STRING | VIT_15s0048g02000.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



