 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00017541-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 332aa MW: 37783 Da PI: 4.4973 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.2 | 3.2e-19 | 49 | 102 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t eq++ Le+ Fe + ++ e++ +LA++lgL+ rqV +WFqNrRa++k
PSME_00017541-RA 49 KKRRLTVEQVRSLERSFEIETKLEPEKKIQLAQELGLQPRQVAIWFQNRRARWK 102
66789************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 131.2 | 3.9e-42 | 49 | 139 | 2 | 92 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
kkrrl+ eqv++LE+sFe e+kLepe+K +la+eLglqprqva+WFqnrRAR+ktkqlE+dy++Lk++yd+lk++ e L++e+e++r+el+
PSME_00017541-RA 49 KKRRLTVEQVRSLERSFEIETKLEPEKKIQLAQELGLQPRQVAIWFQNRRARWKTKQLERDYSVLKASYDTLKSDFEGLQQENENIRAELE 139
9**************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.54E-19 | 40 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.767 | 44 | 104 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.6E-18 | 46 | 108 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.6E-16 | 49 | 102 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.56E-17 | 49 | 105 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.2E-20 | 50 | 111 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.0E-5 | 75 | 84 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 79 | 102 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.0E-5 | 84 | 100 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 5.5E-17 | 104 | 145 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
AFGKFRNNLA MNINDHTYNL SPMANSGNPE EQMDEEAVDE FMNYQPESKK RRLTVEQVRS 60 LERSFEIETK LEPEKKIQLA QELGLQPRQV AIWFQNRRAR WKTKQLERDY SVLKASYDTL 120 KSDFEGLQQE NENIRAELEG LKEKLEEADC SKEDNDSDMG TSTKITSSIE NSKKLAADVV 180 GEETDIDVKP FSDFTLLFKE KDMGSEKMDF KPFRLMLLNE KEGSFSPGSD CSANTRNGDK 240 NPPHQILSQI AQASSSMKYP SGTNNIAAMD SINDQGLMHL AEALCGMGVP SKAYQQLFRI 300 DEGYLHEDAC NHFASEEDQG AASTLPWWEY WP |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 96 | 104 | RRARWKTKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
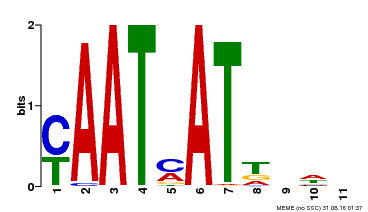
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FJ136970 | 1e-97 | FJ136970.1 Pinus taeda isolate 7088 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136971 | 1e-97 | FJ136971.1 Pinus taeda isolate 7103 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136972 | 1e-97 | FJ136972.1 Pinus taeda isolate 7099 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136973 | 1e-97 | FJ136973.1 Pinus taeda isolate 7087 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136974 | 1e-97 | FJ136974.1 Pinus taeda isolate 7101 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136975 | 1e-97 | FJ136975.1 Pinus taeda isolate 7086 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136976 | 1e-97 | FJ136976.1 Pinus taeda isolate 7093 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136977 | 1e-97 | FJ136977.1 Pinus taeda isolate 7095 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136978 | 1e-97 | FJ136978.1 Pinus taeda isolate 7096 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136979 | 1e-97 | FJ136979.1 Pinus taeda isolate 7089 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136980 | 1e-97 | FJ136980.1 Pinus taeda isolate 7090 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136981 | 1e-97 | FJ136981.1 Pinus taeda isolate 7091 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136982 | 1e-97 | FJ136982.1 Pinus taeda isolate 7100 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136983 | 1e-97 | FJ136983.1 Pinus taeda isolate 7094 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136984 | 1e-97 | FJ136984.1 Pinus taeda isolate 7098 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136985 | 1e-97 | FJ136985.1 Pinus taeda isolate 7102 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136986 | 1e-97 | FJ136986.1 Pinus taeda isolate 7097 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| GenBank | FJ136987 | 1e-97 | FJ136987.1 Pinus taeda isolate 7092 anonymous locus CL3412Contig1_04 genomic sequence. | |||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65310.2 | 2e-38 | homeobox protein 5 | ||||



