 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00017677-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 942aa MW: 103391 Da PI: 6.164 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.9 | 3.6e-18 | 110 | 168 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+++ ++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
PSME_00017677-RA 110 KYVRYTSEQVEALERVYSECPKPSSLRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 168
6789*****************************************************97 PP
| |||||||
| 2 | START | 161.7 | 5.6e-51 | 256 | 464 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galq 92
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+ ++ W ++++ ++l + +g g+++
PSME_00017677-RA 256 IAEETLAEFLSKAKGAAVDWVQMPGMKPGPDSIGIVAISNTCNGVAARACGLVGLDPT-KVAEILKERPSWLRDCRCLDVLTAFPTGngGTIE 347
7899******************************************************.7777777777*****************999**** PP
EEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHH CS
START 93 lmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwll 181
l +++++a ++l++ Rdf+++Ry+ l++g++v++++S++ +q p+ +++vRae+lpSg+li+p+++g+s + +v+h+dl+ ++++++l
PSME_00017677-RA 348 LLYMQTYAATTLASaRDFWTLRYTTVLEDGSLVVCERSLSGTQGGPSippVQHFVRAEMLPSGYLIQPCEGGGSIIRIVDHMDLEPWSVPEVL 440
***********9999*****************************99999999***************************************** PP
HHHHHHHHHHHHHHHHHHTXXXXX CS
START 182 rslvksglaegaktwvatlqrqce 205
r+l++s+++ ++k++ a+l+r+++
PSME_00017677-RA 441 RPLYESSTVLAQKMTIAALRRLRQ 464
*******************99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.775 | 105 | 169 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.1E-14 | 107 | 173 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-16 | 110 | 172 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 8.8E-16 | 110 | 168 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 9.78E-16 | 110 | 170 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-18 | 112 | 168 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 2.05E-6 | 162 | 201 | No hit | No description |
| PROSITE profile | PS50848 | 25.069 | 246 | 461 | IPR002913 | START domain |
| CDD | cd08875 | 3.31E-72 | 250 | 466 | No hit | No description |
| SuperFamily | SSF55961 | 3.43E-35 | 255 | 467 | No hit | No description |
| SMART | SM00234 | 3.5E-36 | 255 | 465 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 5.1E-23 | 256 | 461 | IPR023393 | START-like domain |
| Pfam | PF01852 | 1.7E-48 | 256 | 464 | IPR002913 | START domain |
| Pfam | PF08670 | 7.2E-43 | 791 | 941 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 942 aa Download sequence Send to blast |
MDSYGMPSHV PMHTTQQTQG DNDCAWRHPS TRDDDGNASP YHGIPIISIP CMARIIMTHV 60 MLMGWDGMMR EPRSLAAGVL RAGFESEFRE NLMAVMVNNK DAKNSMDTSK YVRYTSEQVE 120 ALERVYSECP KPSSLRRQQL IRECPILSNI EPKQIKVWFQ NRRCREKQRK EASRLQTVNR 180 KLTAMNKLLM EENDRLQKQV SQLVYENGYM RQQLQNASVA ATDTSCESVV TSGQHQHNPT 240 PQHPPRDASP AGLLSIAEET LAEFLSKAKG AAVDWVQMPG MKPGPDSIGI VAISNTCNGV 300 AARACGLVGL DPTKVAEILK ERPSWLRDCR CLDVLTAFPT GNGGTIELLY MQTYAATTLA 360 SARDFWTLRY TTVLEDGSLV VCERSLSGTQ GGPSIPPVQH FVRAEMLPSG YLIQPCEGGG 420 SIIRIVDHMD LEPWSVPEVL RPLYESSTVL AQKMTIAALR RLRQIAQEAT GEVVFGWGRQ 480 PAVLRTFSQR LSRGFNEAVN GFTDDGWSLM GSDGVEDVTI AINSSPNKHF GSQVNASNGL 540 TTLGGGILCA KASMLLQNVP PALLVRFLRE HRSEWADSNI DAYSAAALKA SPYSVPGSRA 600 GGFSGSQVIL PLAHTVEHEE FLEVIKLEGH GLTQEEAVLS RDMFLLQLCS GIDENAAGAC 660 AELVFAPIDE SFADDAPLLP SGFRVIPLES RTDGSGGPNR TLDLASALEV GSAGTRTSGD 720 SGANSNLRSV LTIAFQFTYE SHLRENVAAM ARQYVRTVVA SVQRVAMALA PSRLSSHVGP 780 RPPPGTPEAL TLARWICQSY RTEFKVLDRL HIGVDLFRAD CDASESVLKL LWHHSDAIMC 840 CSVKSLPVFT FANQAGLDML ETTLVALQDI SLDKILDENG RKSFFTDYGQ IIQQGYAYLP 900 AGICLSSMGR PASYDRAIAW KVLNDEDSTH CIVFMFVNWS FM |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
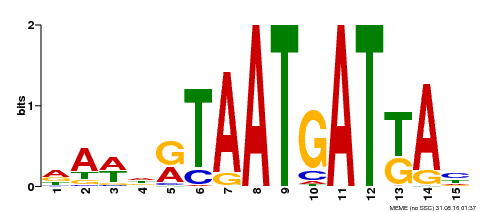
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ385528 | 0.0 | DQ385528.1 Pseudotsuga menziesii class III homeodomain-leucine zipper protein C3HDZ1 (C3HDZ1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010247499.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HOX32-like | ||||
| Swissprot | Q6AST1 | 0.0 | HOX32_ORYSJ; Homeobox-leucine zipper protein HOX32 | ||||
| TrEMBL | Q20BK7 | 0.0 | Q20BK7_PSEMZ; Class III homeodomain-leucine zipper protein C3HDZ1 | ||||
| STRING | XP_010247498.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.2 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



