 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00022003-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 313aa MW: 35049.7 Da PI: 8.0964 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.9 | 1.7e-18 | 172 | 226 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ +++keq Lee F++n+ ++ ++++ LAk+l+L rqV vWFqNrRa+ k
PSME_00022003-RA 172 RKKLRLSKEQSALLEESFKENSSLNPKQKQALAKRLNLRPRQVEVWFQNRRARTK 226
778899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 123.4 | 1.1e-39 | 172 | 261 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
+kk+rlskeq++lLEesF+e++ L+p++K++la++L+l+prqv+vWFqnrRARtk+kq+E+d+e+Lkr++++l++en+rL+ke +eLr +l
PSME_00022003-RA 172 RKKLRLSKEQSALLEESFKENSSLNPKQKQALAKRLNLRPRQVEVWFQNRRARTKLKQTEVDCEFLKRCCESLTDENRRLQKELQELR-AL 261
69*************************************************************************************9.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04618 | 1.7E-9 | 15 | 122 | IPR006712 | HD-ZIP protein, N-terminal |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-17 | 160 | 226 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.18E-18 | 166 | 229 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.491 | 168 | 228 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.9E-16 | 170 | 232 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 8.4E-16 | 172 | 226 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.80E-17 | 172 | 229 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 203 | 226 | IPR017970 | Homeobox, conserved site |
| SMART | SM00340 | 4.5E-26 | 228 | 271 | IPR003106 | Leucine zipper, homeobox-associated |
| Pfam | PF02183 | 8.3E-11 | 228 | 262 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 313 aa Download sequence Send to blast |
MRNYKFLKQM GMDMEDCNTG LGLGMSIGLG MSLIREDLHS HRHHVNGPPV QLDLLPLAPV 60 LQHRDSPWGK SSPGTDGERS AGESKGTMPR RIDVNKLPAS CYYNEDAGTI NVSSPNSALS 120 SFHVDSGGAI NAESSCYAMS VKRERDANEE LEAERACSRV SDEEADQEGG TRKKLRLSKE 180 QSALLEESFK ENSSLNPKQK QALAKRLNLR PRQVEVWFQN RRARTKLKQT EVDCEFLKRC 240 CESLTDENRR LQKELQELRA LKLASPLYMQ MPAATLTMCP SCERVVPAEN SRPPPFTLAK 300 PQFYSYTHSS AAC |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 170 | 176 | TRKKLRL |
| 2 | 220 | 228 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
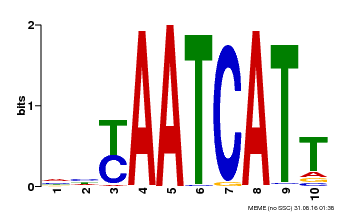
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00477 | DAP | Transfer from AT4G37790 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF081814 | 0.0 | EF081814.1 Picea sitchensis clone WS0294_F04 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018857289.1 | 1e-91 | PREDICTED: homeobox-leucine zipper protein HAT22-like | ||||
| Swissprot | P46604 | 1e-73 | HAT22_ARATH; Homeobox-leucine zipper protein HAT22 | ||||
| Swissprot | P46665 | 3e-73 | HAT14_ARATH; Homeobox-leucine zipper protein HAT14 | ||||
| TrEMBL | A9NKN4 | 0.0 | A9NKN4_PICSI; Uncharacterized protein | ||||
| STRING | XP_010248307.1 | 1e-89 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37790.1 | 1e-75 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



