 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00022730-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 872aa MW: 96862.7 Da PI: 5.8971 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 75 | 8.9e-24 | 167 | 268 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++ +++a+e+ +++++ s++l+ +d++g +W++++i+r++++r++lt+GW+ Fv++++L +gD ++F + +
PSME_00022730-RA 167 FCKTLTASDTSTHGGFSVLRRHADEClpsldMSQQPPSQELVAKDLHGVEWRFRHIFRGQPRRHLLTTGWSVFVSSKRLVAGDAFIFL--RGE 257
99*****************************55566678*************************************************..449 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
++el+v+v+r+
PSME_00022730-RA 258 NGELRVGVRRA 268
999*****996 PP
| |||||||
| 2 | Auxin_resp | 123.6 | 1.4e-40 | 293 | 375 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+st+++F+v+Y+Pr+s+seF+++++++++a+k+++svGmRfkm+fe+e+++e+r++Gt+vg+ d+dpv+Wp+SkWr+Lk
PSME_00022730-RA 293 ASHAVSTRTMFSVYYKPRTSPSEFIIPYDQYMEAMKTNFSVGMRFKMKFEGEEVPEQRFTGTIVGICDVDPVSWPGSKWRCLK 375
789******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 6.8E-42 | 160 | 282 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.09E-43 | 161 | 296 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.19E-21 | 166 | 267 | No hit | No description |
| Pfam | PF02362 | 1.2E-21 | 167 | 268 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.0E-23 | 167 | 269 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.012 | 167 | 269 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.2E-38 | 293 | 375 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 23.564 | 757 | 842 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 5.89E-9 | 759 | 830 | No hit | No description |
| Pfam | PF02309 | 6.1E-5 | 802 | 846 | IPR033389 | AUX/IAA domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 872 aa Download sequence Send to blast |
MAYAENFRNF TCRIPASNQS NSSNGGVDGY SSMPNEGGLV LGQIGGSHGY LNSSQSVNDA 60 LYEELWHACA GPLVTLPRIG ERVFYFPQGH MEQVEASTNQ GADQHMPLFN LPFKILCRVI 120 NVQLKAEADT DEVFSQITLV PEVEQDESSV DKEPLPPLAP KPLVYSFCKT LTASDTSTHG 180 GFSVLRRHAD ECLPSLDMSQ QPPSQELVAK DLHGVEWRFR HIFRGQPRRH LLTTGWSVFV 240 SSKRLVAGDA FIFLRGENGE LRVGVRRAMR QQNNVPSSVI SSHSMHLGVI ATASHAVSTR 300 TMFSVYYKPR TSPSEFIIPY DQYMEAMKTN FSVGMRFKMK FEGEEVPEQR FTGTIVGICD 360 VDPVSWPGSK WRCLKVQWDE ISAITRPERV SPWKLEPSLA PASVNSLPVA RGKRPRPNIL 420 PLSSDLSVLD KASVDSTQVH RFPRVLQGQE VMTLGGSLGD SDLESGQKTV VWGSKLDDVK 480 SEGMGCQRRL LSENWMPPLR HDSVYSDAFS SFQPMGEMQE FRGSLPNSIL EDAQQPKVSR 540 KQFQDQEGKI VDASGLWSTN FPNSLQEMSV TSAAQSHRQS GNARWPGLNG HPQFSNIGME 600 SLVGNWQMHM ISQCQSEASV AHGGADRNAA RTLQVNNMGG TEPQPSFDLW ETSKGAEVSR 660 PTDSNCKLFG FHLIDNSTGG ELNLSKIRGS VNEEDLQVAA HASTEIRSQS VEPEQQSEQT 720 KTTKSDVPAA SCEQEKTSPR SSNETQCRAS AQSNPMRSCT KVQKQGSALG RAVDLTKFEG 780 YTELIRELEL MFNFEGQLQD ASKGWQVVYT DDEGDMMLVG DDPWHREFCC MVRKIFIYTR 840 EEVEKMTPSA APNMKIPGHS EEEKVARETP TQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-179 | 57 | 397 | 15 | 354 | Auxin response factor 1 |
| 4ldw_A | 1e-179 | 57 | 397 | 15 | 354 | Auxin response factor 1 |
| 4ldw_B | 1e-179 | 57 | 397 | 15 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 412 | 416 | KRPRP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Could act as transcriptional activator or repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Functions independently of ethylene and cytokinin response pathways. May act as a repressor of cell division and organ growth. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952, ECO:0000269|PubMed:16339187}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
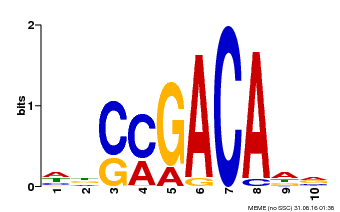
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT116582 | 0.0 | BT116582.1 Picea glauca clone GQ03802_H23 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010942378.1 | 0.0 | auxin response factor 23 | ||||
| Swissprot | Q94JM3 | 0.0 | ARFB_ARATH; Auxin response factor 2 | ||||
| TrEMBL | A0A0C9RYI4 | 0.0 | A0A0C9RYI4_9SPER; Auxin response factor | ||||
| STRING | XP_008776920.1 | 0.0 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||



