 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00038811-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 813aa MW: 86570.1 Da PI: 7.2499 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.3 | 8.6e-16 | 552 | 598 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
PSME_00038811-RA 552 VHNLSERRRRDRINEKMRALQELIPHC-----NKSDKASMLDEAIEYLKTLQ 598
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.44E-16 | 544 | 602 | No hit | No description |
| SuperFamily | SSF47459 | 1.11E-19 | 545 | 606 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 7.4E-20 | 545 | 606 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.854 | 548 | 597 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.0E-13 | 552 | 598 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.6E-17 | 554 | 603 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 813 aa Download sequence Send to blast |
MPWSGRNSTP VRPTAGNEQG GLCSRIADKS YKEPPGPNEY LLSQQIDLKD SMNHCVPNWD 60 IENGMMQSSD FLSMAKANWD NVQPPKRSMM LEHDIEELLW ENGQVVNKVV KRPLPTQLSK 120 TLQNDDAVLP GKPSPVGKDN TMELVVHDMS AANASSIHED VMDSWLHYPL DDSLEKEYCS 180 DFFGGMPSTN VHMLRDTLSG QMNNVPAEKV SNLHVQKESS LSSESNSAPW TGVFAGTVNT 240 TNAAGPSKTG KESPSKSISS DKAMALGAGR ASGTIPQSVT DAFAKVRTVS QLPVSKWHTD 300 VQDSSRSKDD HLACKKTATQ VSVSPLSMPP PKIQATDLAS MKPSRSKIVN FSHFSRPAAT 360 MKANLQSIGS ASGLSTTGIQ NRLDKIRMDG NAAAEPSIIE SNSTGMTTIG SSSGANSRAQ 420 DIGSCQTHQN PPSGKNLDVA TCSEDVTDSS AKASEQGVCQ NSNAGRSLAS CGTEKCGDAG 480 DGPEPTITSS SGGSGYSAGR VGKEATNTSK RKGRDVEDSE CQSEDVEYES ADTKKQGPSR 540 TTTSKRSRAA EVHNLSERRR RDRINEKMRA LQELIPHCNK SDKASMLDEA IEYLKTLQLQ 600 VQIMSMGGGM GMVFPGGMQH FQVPQMAHMS PMGMGIGMGY SMGMLDMAAT SGRPVMSLPS 660 MHVSALPGSA IHCQAALPLS GMPGPSLPMS RLPGPGLQVS GLPVSTLPVS GLHGTTQPGL 720 VSTSAPGSTD LQDHMQNANV MGHYKHYMNN HHQMQGPPQV INGNLYNASM AQPPPQNPVQ 780 SSGRGIHNAA IASGKTGTTG KHYDCYCSFV LGK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 556 | 561 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
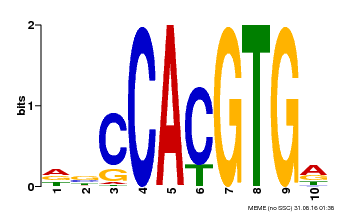
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT123569 | 0.0 | BT123569.1 Picea sitchensis clone WS04714_N07 unknown mRNA. | |||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 7e-39 | phytochrome interacting factor 3 | ||||



