 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00039531-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 734aa MW: 81268.4 Da PI: 6.3575 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 34.9 | 3.9e-11 | 358 | 414 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng.krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g rk + g ++ +e+Aak ++ a++k++g
PSME_00039531-RA 358 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqTRKGRQ-GGYDKEEKAAKSYDLAALKYWG 414
78*******************77776664446655.779999**************98 PP
| |||||||
| 2 | AP2 | 46 | 1.3e-14 | 459 | 508 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GV+++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
PSME_00039531-RA 459 YRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 508
9***************988532...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 8.92E-18 | 358 | 422 | No hit | No description |
| SuperFamily | SSF54171 | 3.27E-13 | 358 | 424 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 6.4E-8 | 358 | 414 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.898 | 359 | 422 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.3E-22 | 359 | 428 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.7E-11 | 359 | 423 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.1E-6 | 360 | 371 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.94E-25 | 457 | 518 | No hit | No description |
| SuperFamily | SSF54171 | 1.05E-17 | 457 | 518 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 19.23 | 458 | 516 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.9E-34 | 458 | 522 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-18 | 458 | 516 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.5E-9 | 459 | 508 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.1E-6 | 498 | 518 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 734 aa Download sequence Send to blast |
MKYMNNWLAF SLSPHLAIDV SASADQSAQP HQLSSVPTSF NCLQTSQMPR STLPYQSYGV 60 NSENGGNSFY APMPLKSDGS LCIMEALGRS QIEEWHDLKP PGLQAGRVQS PNVMGSRLSS 120 SDSQYEITPN GASNHGSPKL EDFLGGASLG SHEYSASARD HQSMSLSLQN SIYFDHNVGE 180 DNNGSCKMIN VNQMPYSECR RMGVPSSFHT YGDNGNNTYE AAMHDQGLSQ PNMFADCSLQ 240 FNPPGPGIIE NPSSCMVGIS AMKTLLRQYP NGGSSDKNST NESHETLNNI GDLQSQALTL 300 TMSPGSQSSS VTIVPHSGTN TECVAVETSR KRVAGSKSGN TRQPVPRKSI DTFGQRTSQY 360 RGVTRHRWTG RYEAHLWDNS CKKEGQTRKG RQGGYDKEEK AAKSYDLAAL KYWGPSTHIN 420 FPLNTYEKEL EEMKHMTRQE YVANLRRKSS GFSRGASMYR GVTRHHQHGR WQARIGRVAG 480 NKDLYLGTFS TQEEAAEAYD IAAIKFRGTN AVTNFDISRY DVKRILASNT LLVGELARLN 540 RDQLAEPLTA EPDQVDVPIV IHQIANEKYN AKETDYEDED TKNNWQMQMS GDQEQNQINS 600 CSNEASDHDK MWDDQKPSQS FTESQHLAAL HNLMGLDHAE RNEDSSHNNK NNSLLSDMSA 660 SDPRSPGASM GREEHEEEAA LSPSGQNENG LFFSSKQPAL QINNALSSWM TQAARPALNS 720 MAHHIPMFAS WNDT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as positive regulator of adventitious (crown) root formation by promoting its initiation. Promotes adventitious root initiation through repression of cytokinin signaling by positively regulating the two-component response regulator RR1. Regulated by the auxin response factor and transcriptional activator ARF23/ARF1. {ECO:0000269|PubMed:21481033}. | |||||
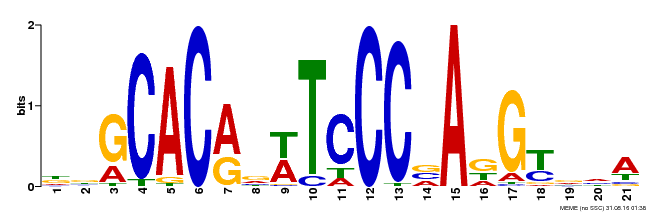
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by auxin. {ECO:0000269|PubMed:21481033}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT100555 | 1e-127 | BT100555.1 Picea glauca clone GQ0025_E20 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Swissprot | Q84Z02 | 1e-123 | CRL5_ORYSJ; AP2-like ethylene-responsive transcription factor CRL5 | ||||
| TrEMBL | Q2Z2G4 | 0.0 | Q2Z2G4_GINBI; AINTEGUMENTA-like protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-119 | AP2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



