 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00042339-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 312aa MW: 35012.4 Da PI: 9.0567 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 28 | 4.9e-09 | 36 | 80 | 4 | 46 |
S-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 4 WTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwqk 46
W+ + +l+ +a + + ++ +W+++a++++ g+t+ +++ ++ +
PSME_00042339-RA 36 WSLQQNKLFENALAIYDKDtpdRWEKVAKMLP-GKTAIDVRKHYED 80
********************************.***********76 PP
| |||||||
| 2 | Myb_DNA-binding | 42.4 | 1.6e-13 | 143 | 187 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ +++ + +++G+g+W++I+r + Rt+ q+ s+ qky
PSME_00042339-RA 143 PWTEDEHRQFLLGLHKFGKGDWRSISRNFVVSRTPTQVASHAQKY 187
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 6.597 | 31 | 86 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.5E-8 | 32 | 84 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 7.59E-12 | 34 | 90 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-4 | 36 | 82 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.5E-6 | 36 | 81 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.81E-6 | 42 | 82 | No hit | No description |
| PROSITE profile | PS51294 | 18.439 | 136 | 192 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.45E-16 | 137 | 192 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.8E-18 | 139 | 190 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 6.4E-11 | 140 | 186 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.7E-11 | 140 | 190 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.96E-10 | 143 | 188 | No hit | No description |
| Pfam | PF00249 | 2.2E-11 | 143 | 187 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MPYFLSSFSQ DFKICAMWEM EFPSQETSLS SSGSFWSLQQ NKLFENALAI YDKDTPDRWE 60 KVAKMLPGKT AIDVRKHYED LVEDVTCIED GRVALPTYSN SSCTLEWIAE KSGDMHGLKQ 120 QFGSGGKGPS TKPSEQERKK GVPWTEDEHR QFLLGLHKFG KGDWRSISRN FVVSRTPTQV 180 ASHAQKYFIR LSSDNKNKRR SSIHDITTVN GTDRRPFPLP QTSTRQTNSP LRQADINHSP 240 CLTYPSPISR GPLINSLGSQ ADGSPLMSPH YPLSLYTQRG FGGHSPRIII PDSSIGIGMP 300 HLGYPGQPAM QY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 2e-20 | 28 | 101 | 3 | 76 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
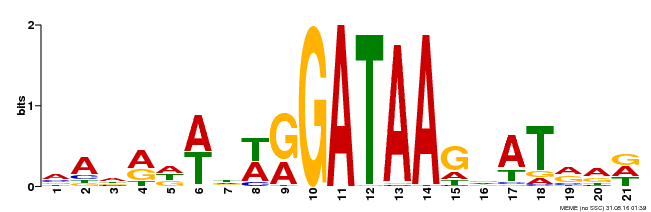
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT107022 | 0.0 | BT107022.1 Picea glauca clone GQ03101_N16 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006429310.1 | 4e-81 | transcription factor DIVARICATA | ||||
| Refseq | XP_006480971.1 | 4e-81 | transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 2e-73 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A0D6R2Q1 | 4e-90 | A0A0D6R2Q1_ARACU; Uncharacterized protein | ||||
| STRING | XP_006480971.1 | 1e-80 | (Citrus sinensis) | ||||
| STRING | XP_006429310.1 | 2e-80 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38090.1 | 2e-69 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



