 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00043147-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 647aa MW: 70111.3 Da PI: 7.52 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 95.5 | 3.8e-30 | 371 | 429 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
+Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+aed +v+++tYeg+Hnh+
PSME_00043147-RA 371 SDGCQWRKYGQKMAKGNPCPRAYYRCTMAvGCPVRKQVQRCAEDRTVLVTTYEGSHNHP 429
7*********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 6.9E-33 | 355 | 431 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.96E-27 | 363 | 431 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.7 | 365 | 431 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.8E-36 | 370 | 430 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.2E-26 | 372 | 429 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0080169 | Biological Process | cellular response to boron-containing substance deprivation | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 647 aa Download sequence Send to blast |
MEAVGLSIAG NGSYRELLDT FLKQKCRRKT DFMADARRFD KLGGIDLSVK LEETENIAEE 60 KLMRQREPEI EIKNGVVDFF SEKSSRGLGF EFRPTTGGVA PVLSSLGEGL RLGSQKSENK 120 DEGPSSSKGI SFSTGLQLLP ANTASDRSTV DDGMSPVRSS EPSNPEEAEP KSEVELNRMN 180 EENQRLRFML NYISKNYGNL QMHLMSVMQQ QPPSDKVEEK FNTSSAKSQD GVKVDAVAPV 240 QRQFLDLVPI SSHGTDSQHS LSSESLDDID QEAAAKSNNI EVLSKKRDLC TANSTDKARL 300 QQASSDRAAS PNRLNGQGQE KGKPDSPPDH DSWTANKAQK VPNKCAVDQT EATIRKARVS 360 VRARSEASMI SDGCQWRKYG QKMAKGNPCP RAYYRCTMAV GCPVRKQVQR CAEDRTVLVT 420 TYEGSHNHPL PPAATAMAST TSAAACMLLS GSTTSNDGGF NSSFMQAAAG SLMSCSPASM 480 ATISASAPFP TVTLDLTQNP NHNNMYQRPP SGFPMPLGGF QQFPSHPPQI FSHSLLNNSK 540 FAPLSGQQFD PQQQLPPYSG HPMQQQQQQQ QQQQTHMPVP ISQFKGPQSS LIDTVSAITA 600 DPNFTSALAA AITSIISGQP NNANSIPSQA LPGTARADNP ASAKLHQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 1e-21 | 357 | 433 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
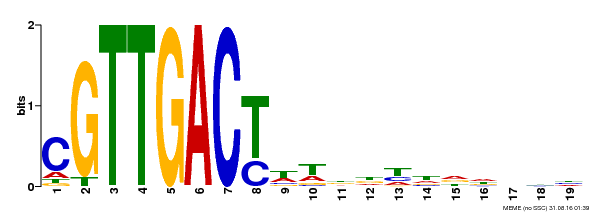
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00209 | DAP | Transfer from AT1G62300 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT114529 | 0.0 | BT114529.1 Picea glauca clone GQ03516_F03 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010252466.1 | 1e-131 | PREDICTED: probable WRKY transcription factor 31 | ||||
| TrEMBL | A0A0C9QVA3 | 0.0 | A0A0C9QVA3_9SPER; TSA: Wollemia nobilis Ref_Wollemi_Transcript_6922_2664 transcribed RNA sequence | ||||
| STRING | XP_010252466.1 | 1e-131 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62300.1 | 2e-81 | WRKY family protein | ||||



