 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00047903-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 805aa MW: 89186.3 Da PI: 8.7214 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.8 | 2.1e-23 | 159 | 260 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldg 86
f k+lt+sd+++ g +++p+ +ae++ + ++ + t+ +d +g++W++++iyr++++r++lt+GW+ Fv++++L +gD +vF +
PSME_00047903-RA 159 FAKTLTQSDANNGGGFSVPRFCAETIfprldysSDPPVQ--TVLAKDVHGEVWKFRHIYRGTPRRHLLTTGWSTFVNQKKLVAGDAIVFL--R 247
89*********************999****995444444..8999*********************************************..4 PP
SSEE..EEEEE-S CS
B3 87 rsefelvvkvfrk 99
+ ++el+v+v+r+
PSME_00047903-RA 248 SATGELWVGVRRS 260
479999*****97 PP
| |||||||
| 2 | Auxin_resp | 105.5 | 6.1e-35 | 338 | 419 | 3 | 83 |
Auxin_resp 3 haastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrl.sGtvvgvsdldpvrWpnSkWrsLk 83
+ a ++++FevvY+Prast+eF+vk+++v +al++++s+GmRfkm fetedss++++ +Gtv+ v+ +dpv WpnS+Wr+L+
PSME_00047903-RA 338 TLAVSGQPFEVVYYPRASTPEFCVKAKSVDSALRIQWSPGMRFKMPFETEDSSRISWfMGTVSFVQFADPVLWPNSPWRLLQ 419
677899**************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.49E-29 | 153 | 285 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.8E-35 | 154 | 270 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.31E-20 | 158 | 259 | No hit | No description |
| PROSITE profile | PS50863 | 12.52 | 159 | 261 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.6E-19 | 159 | 261 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.7E-22 | 159 | 260 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.8E-30 | 337 | 419 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 16.57 | 719 | 799 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007389 | Biological Process | pattern specification process | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0048829 | Biological Process | root cap development | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 805 aa Download sequence Send to blast |
MERGLRFAAC CRSVYSPCAR TMLRKMESGE VKIEKGLDPQ LWHACAGGMV QMPAINSKVY 60 YYPQGHAEHA ATVPDFAGNM RMSPSILCRV VSVKFLADTE TDEVYARMRL IPISDDEQNS 120 SVEDSEAESR PPSPPRTPVA PNSQTQTENG NKNHPPASFA KTLTQSDANN GGGFSVPRFC 180 AETIFPRLDY SSDPPVQTVL AKDVHGEVWK FRHIYRGTPR RHLLTTGWST FVNQKKLVAG 240 DAIVFLRSAT GELWVGVRRS TRSTTESHSH SHLQSSSAWH LPVSSITASP PPPYRASRWE 300 VKAADYFSGI ATANTNGSTG FSRNRGRVSA KSVIEACTLA VSGQPFEVVY YPRASTPEFC 360 VKAKSVDSAL RIQWSPGMRF KMPFETEDSS RISWFMGTVS FVQFADPVLW PNSPWRLLQV 420 TWDEPDMLQN VKRVSPWLVE LVSNMPPIQM TPFTLPKKKL RVTQSPDFQL EGQGIMGLQM 480 STVSNSLLRQ INPWHGISDN VPVGIQGARQ DRIYGISLSD IFPNKGQNGL FFKHPSHQEQ 540 GVMPTRVCTE LNIGNSPQHD GSGMQSSPSS FLTVGNSSQN EQKTSSGSAS SSTTKGSSFL 600 LFGKPIHTAP SPKSCQQQQS GLSSSDGTAF HMFSENGSSR LTSNNSSNGN QEALERVQKL 660 VTGRRDGTKL SENYGLRLSH GESLDYAANG MRSLQWFKDQ TSVLKLEKDK QENTFEDSIV 720 HCKVFKETEE VGRTLDLSLF CTYEQLYDKL AKMFGIDESE LSNRVLYKGL GGSVRHAGDE 780 PYKDFMKTVR RLTILSDSGS DNMAR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldy_A | 5e-69 | 33 | 440 | 15 | 353 | Auxin response factor 1 |
| 4ldy_B | 5e-69 | 33 | 440 | 15 | 353 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00461 | DAP | Transfer from AT4G30080 | Download |
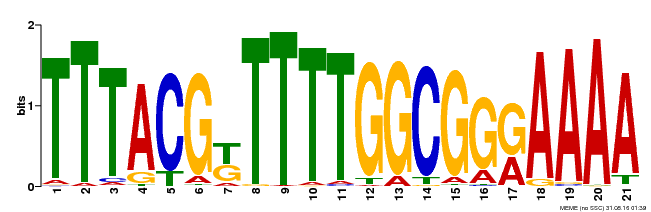
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT119496 | 0.0 | BT119496.1 Picea glauca clone GQ04106_G13 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011624034.1 | 0.0 | auxin response factor 18 | ||||
| TrEMBL | A0A0D6QZZ4 | 0.0 | A0A0D6QZZ4_ARACU; Auxin response factor | ||||
| STRING | ERN07701 | 0.0 | (Amborella trichopoda) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G30080.1 | 0.0 | auxin response factor 16 | ||||



