 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00051212-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 472aa MW: 52540.4 Da PI: 5.8399 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 87.4 | 1.2e-27 | 188 | 248 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK++ g+++prsYYrCt++ gC+++kkv+r +++p+v+eitY+g H ++
PSME_00051212-RA 188 EDGYTWRKYGQKDILGAQHPRSYYRCTHKhdlGCQATKKVQRPDDNPSVFEITYSGVHSCQ 248
7***************************99999*************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.1E-26 | 185 | 248 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.88E-24 | 186 | 248 | IPR003657 | WRKY domain |
| SMART | SM00774 | 5.8E-38 | 187 | 249 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.0E-26 | 188 | 248 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 20.541 | 188 | 250 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010193 | Biological Process | response to ozone | ||||
| GO:0042542 | Biological Process | response to hydrogen peroxide | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 472 aa Download sequence Send to blast |
MADTRDVKDF DECLKEVNHG CQMAKELETK LNSAKGPDLF ELLWASSAEI VATFSHALEM 60 LRSNSSTPSC LDNWQEELKA ILEKQLGGLE VVHSHKKSVK DHLNSSPRSG GRSSLSGSPP 120 SDMSEGSPAR SLATPSKLKR NDLADTSENE RASNKERRGI SRKRKSLPKW IKIMPANSSE 180 PGNDIPPEDG YTWRKYGQKD ILGAQHPRSY YRCTHKHDLG CQATKKVQRP DDNPSVFEIT 240 YSGVHSCQSD ARLLHFPFPI SQIQGSNIIP NSRTPTTLVF GTDESNLQSN PLFPPTPFLS 300 SSATPWFPAD DGGVCQVKSE SLDSFGKSPS YSPNIDSSTH NAMQPFMTSP TADPIYRLRD 360 SEKEFSADFI PVHHQHNEYP STFPGGELEF PNFAPFFCSP CTSESNYMPP PPPLMQAMDT 420 SRQRSQASES DFTEVVSTHN SATDSPMLEM DHFPTMFTMV NLDSMFERSD NF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 1e-19 | 188 | 248 | 9 | 69 | OsWRKY45 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 160 | 165 | SRKRKS |
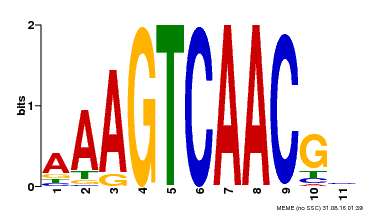
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00525 | DAP | Transfer from AT5G24110 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT116253 | 0.0 | BT116253.1 Picea glauca clone GQ03714_K22 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0C9S9Q1 | 1e-100 | A0A0C9S9Q1_9SPER; TSA: Wollemia nobilis Ref_Wollemi_Transcript_7171_2278 transcribed RNA sequence | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G11070.2 | 7e-29 | WRKY family protein | ||||



