 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PSME_00056295-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Pseudotsuga
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 195aa MW: 22259.3 Da PI: 6.768 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 85.3 | 3.6e-27 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien + rq tfskRr g+lKKA ELS+LCdae+av+ifss+gklye++s
PSME_00056295-RA 9 KRIENPTSRQATFSKRRGGLLKKARELSILCDAEIAVLIFSSSGKLYEFAS 59
79***********************************************86 PP
| |||||||
| 2 | K-box | 61.1 | 4.4e-21 | 86 | 178 | 8 | 99 |
K-box 8 s.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkle 99
+ e++++++ + ++akL+ + e Lq ++h GedLe L+ +Lq Le++Le ++ ++R++Kn+ +l+ i+e + e++l een++Lr+k++
PSME_00056295-RA 86 DcSENNQNNNQNIQAAKLRIQAEHLQLMLKHTSGEDLEGLDTDQLQTLEENLELGIGRVRARKNQCMLSRIKEISNIEQDLIEENRTLRRKMN 178
3233344444445789**************************************************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 32.436 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 1.9E-40 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 5.27E-43 | 2 | 78 | No hit | No description |
| SuperFamily | SSF55455 | 9.55E-33 | 2 | 76 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.2E-29 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 2.1E-25 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.2E-29 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.2E-29 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.6E-17 | 89 | 177 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 12.445 | 93 | 183 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010440 | Biological Process | stomatal lineage progression | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008134 | Molecular Function | transcription factor binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 195 aa Download sequence Send to blast |
MGRGKIEIKR IENPTSRQAT FSKRRGGLLK KARELSILCD AEIAVLIFSS SGKLYEFAST 60 EMKTILDRYS KCFEQSSQSE SQALVDCSEN NQNNNQNIQA AKLRIQAEHL QLMLKHTSGE 120 DLEGLDTDQL QTLEENLELG IGRVRARKNQ CMLSRIKEIS NIEQDLIEEN RTLRRKMNDF 180 LNNECLTAVP HTLET |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6byy_A | 8e-23 | 1 | 89 | 1 | 86 | MEF2 CHIMERA |
| 6byy_B | 8e-23 | 1 | 89 | 1 | 86 | MEF2 CHIMERA |
| 6byy_C | 8e-23 | 1 | 89 | 1 | 86 | MEF2 CHIMERA |
| 6byy_D | 8e-23 | 1 | 89 | 1 | 86 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 141 | 149 | GRVRARKNQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00410 | DAP | Transfer from AT3G57230 | Download |
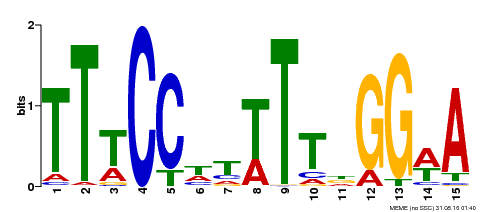
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001266821.1 | 2e-49 | uncharacterized protein LOC100832541 | ||||
| Refseq | XP_010230319.1 | 2e-49 | uncharacterized protein LOC100832541 isoform X1 | ||||
| Refseq | XP_024313924.1 | 2e-49 | uncharacterized protein LOC100832541 isoform X1 | ||||
| Refseq | XP_024538443.1 | 2e-49 | MADS-box transcription factor 27 isoform X3 | ||||
| Refseq | XP_024540272.1 | 2e-49 | MADS-box transcription factor 27 isoform X7 | ||||
| Swissprot | Q6VAM4 | 6e-46 | MAD23_ORYSJ; MADS-box transcription factor 23 | ||||
| TrEMBL | Q8LLC8 | 5e-50 | Q8LLC8_SPIAN; MADS-box gene 2 protein | ||||
| STRING | BRADI2G25090.1 | 7e-49 | (Brachypodium distachyon) | ||||
| STRING | PP1S209_130V6.2 | 2e-48 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G57230.1 | 1e-45 | AGAMOUS-like 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



