 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.A00033.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 864aa MW: 97492.9 Da PI: 7.5877 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12 | 0.00065 | 557 | 579 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C Cg +F + +Lk+H+++H
Pahal.A00033.1 557 HRCLQCGACFQKPAHLKQHMQSH 579
79*******************99 PP
| |||||||
| 2 | zf-C2H2 | 22.4 | 3.1e-07 | 585 | 609 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp dC+ s++rk++L+rH+ +H
Pahal.A00033.1 585 FICPleDCPFSYKRKDHLNRHMLKH 609
78*********************99 PP
| |||||||
| 3 | zf-C2H2 | 11.3 | 0.0011 | 616 | 639 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirt.H 23
C+ C+++Fs k n++rH + H
Pahal.A00033.1 616 CTvdGCDRRFSIKANMQRHVKEfH 639
88889***************9988 PP
| |||||||
| 4 | zf-C2H2 | 13.3 | 0.00024 | 651 | 675 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C+ C+k F+ s Lk+H +H
Pahal.A00033.1 651 FICKeeGCNKAFKYFSKLKKHEESH 675
789999***************9988 PP
| |||||||
| 5 | zf-C2H2 | 17.8 | 9.4e-06 | 742 | 766 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C+ sFs+ksnL++H++ H
Pahal.A00033.1 742 KCTfdGCDHSFSNKSNLTKHMKAcH 766
799999****************955 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.25.40.10 | 5.4E-7 | 32 | 163 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 5.196 | 35 | 65 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.934 | 66 | 100 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.11 | 68 | 97 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 7.903 | 136 | 166 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.0033 | 141 | 163 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.745 | 167 | 201 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.018 | 169 | 197 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.01 | 200 | 226 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 5.733 | 202 | 232 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 5.4E-7 | 250 | 326 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 8.659 | 268 | 298 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 0.0012 | 273 | 298 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF13041 | 6.5E-11 | 298 | 346 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 12.09 | 299 | 333 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 3.5E-7 | 301 | 335 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.988 | 334 | 364 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 6.686 | 370 | 400 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.018 | 373 | 397 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 5.4E-7 | 387 | 464 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 7.52 | 436 | 470 | IPR002885 | Pentatricopeptide repeat |
| SMART | SM00355 | 22 | 511 | 533 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 7.3E-4 | 554 | 576 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 12.383 | 557 | 584 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.057 | 557 | 579 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 559 | 579 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.88E-10 | 569 | 611 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-11 | 577 | 611 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.037 | 585 | 614 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 1.9E-4 | 585 | 609 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 587 | 609 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.66E-7 | 595 | 645 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.2E-8 | 612 | 641 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.512 | 614 | 644 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.027 | 614 | 639 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 616 | 639 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.2E-6 | 648 | 675 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.18 | 651 | 675 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.013 | 651 | 675 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 653 | 675 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 7.5 | 683 | 708 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 685 | 708 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 24 | 711 | 732 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 3.15E-9 | 726 | 783 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-6 | 726 | 763 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.884 | 741 | 771 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.1E-4 | 741 | 766 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 743 | 766 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.1E-5 | 770 | 794 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.245 | 772 | 798 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.6 | 772 | 798 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 774 | 798 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 864 aa Download sequence Send to blast |
MNRSICHHLL SQCKTLRELQ KLHAQAVAQG LHPHRQSVSC KIFRCYADFG RAADARKLFD 60 EIPRPDLVSF TSLMSLHIQL ERHREAVSLF SLAVSDGHRP DGFAVVGALS ASGGAGDQQV 120 GRAVHGLIFR LGLDSEVVVG NALVDMYSRC GKFESAVLVF DRMFLKDEIT WGSMLHGYIK 180 CAGLDLALSF FDQMPVKSVV AWTALITSHV QGRLPVRALE LFARMILESH RPTHVTIVGV 240 LSACADIGAL DLGRIIHGYG SKCNASSNII VSNALMDMYA KSGSIEMALS VFQEVQSKDA 300 FTWTTMISCF TVQGDGRKAL ELFWDMLRSG VVPNSVTFVS VLSACSHAGL IEEGRELFGT 360 MRETYSIDPQ IEHYGCMIDL LGRGGLLEEA EALIVDMNVE PDIVIWRSLL SACLVHHNDR 420 LAEIAGKEIM KREPGDDGVY VLLWNMYASS NKWREAREIR QQMLTRKIFK KPGCSWIEVD 480 GVVHEFLMCS GDEIDGDASV GQKGFKDIRR YKCEFCTVVR SKKCLIQAHM VAHHKDELDK 540 SETYDSNGEK IVCEEEHRCL QCGACFQKPA HLKQHMQSHS NERLFICPLE DCPFSYKRKD 600 HLNRHMLKHQ GKLFGCTVDG CDRRFSIKAN MQRHVKEFHE DENVTKSNQQ FICKEEGCNK 660 AFKYFSKLKK HEESHVKLNY IEVVCCEPGC MKMFTNVDCL RAHNQSCHQH VQCEMCGEKH 720 LKKNIKRHLQ AHDEVPSGER IKCTFDGCDH SFSNKSNLTK HMKACHDQLK PFTCRVAGCG 780 KAFTYKHVRD NHEKSSAHVY VEGDFEEMDE QLRSRPRGGR KRKALTVETL TRKRVTILGE 840 ASSLDDGEEY LRWLLSSGDD SAQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5iww_D | 4e-56 | 9 | 413 | 3 | 338 | PLS9-PPR |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 814 | 822 | RPRGGRKRK |
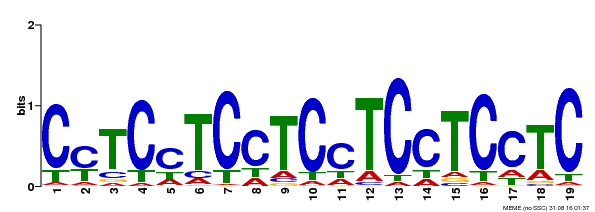
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795590.1 | 0.0 | pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like | ||||
| TrEMBL | K3Z0X6 | 0.0 | K3Z0X6_SETIT; Uncharacterized protein | ||||
| STRING | Pavir.J03392.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-116 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.A00033.1 |



