 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.A03010.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 272aa MW: 27998.6 Da PI: 3.6974 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 30.3 | 9.6e-10 | 2 | 29 | 19 | 48 |
TTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 19 lGggtWktIartmgkgRtlkqcksrwqkyl 48
lG++ W++Ia++++ gRt++++k+ w+++l
Pahal.A03010.2 2 LGNK-WSKIAACLP-GRTDNEIKNVWNTHL 29
8999.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00249 | 1.2E-7 | 1 | 29 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 16.959 | 1 | 33 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-13 | 2 | 30 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.04E-8 | 2 | 33 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.30E-6 | 4 | 29 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MLGNKWSKIA ACLPGRTDNE IKNVWNTHLK KKAALREQQK AGAAKNDGAA SGGDAGTPVT 60 DSSSASSSTT TTNSSSGGSD SGDQCDTSKE PDAVPVSPLQ LEDVDVSEML VDAPAAAQPM 120 LSASCSSSSL TTCAGGVEDL IELPVIDIEP DIWSIIDGEC ADASGARRGD ATAPCTGAAV 180 STSEAGEAAN DWWLENLEKE LGLWGPAEDP QAQTDLLDHT GFGPLVDSEG DPVSTYFQTG 240 PDNAAAELLL DVELELELEL EPEPEPSALL L* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that regulates positively genes involved in anthocyanin biosynthesis such as A1. {ECO:0000269|PubMed:7920701}. | |||||
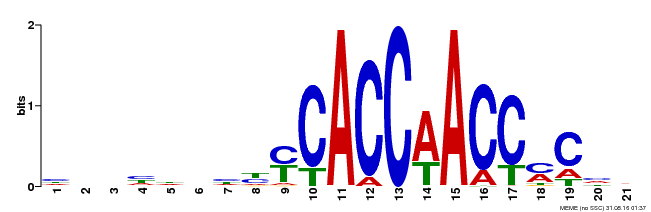
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00249 | DAP | Transfer from AT1G79180 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025814304.1 | 0.0 | myb-related protein Zm1-like | ||||
| Swissprot | P20024 | 1e-101 | MYB1_MAIZE; Myb-related protein Zm1 | ||||
| TrEMBL | A0A2S3GS79 | 0.0 | A0A2S3GS79_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Aa01159.1.p | 1e-115 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G16490.1 | 4e-16 | myb domain protein 58 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.A03010.2 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



