 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.A03051.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 276aa MW: 30254.3 Da PI: 8.6307 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 107.1 | 9.4e-34 | 26 | 80 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWtp+LHerFveav++LGG++kAtPk++l+lm++kgLtl+h+kSHLQkYRl
Pahal.A03051.1 26 KPRLRWTPDLHERFVEAVTKLGGPDKATPKSVLRLMGMKGLTLYHLKSHLQKYRL 80
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.032 | 23 | 83 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.6E-33 | 23 | 81 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.85E-17 | 26 | 82 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.6E-25 | 26 | 81 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.4E-10 | 28 | 79 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.7E-24 | 125 | 169 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
MFEGMERAGY GGAAAMGGVV LSRDPKPRLR WTPDLHERFV EAVTKLGGPD KATPKSVLRL 60 MGMKGLTLYH LKSHLQKYRL GKQSKKDTGL EASRGAFAAQ GINFSAPVPP SIPSTAGNNT 120 GETPLADALK YQIEVQRKLH EQLEVQKKLQ MRIEAQGKYL QTILEKAQNN LSYDATGAAN 180 LEATRSQLTD FNLALSGFMD NVSQACEQNN GELAKAMTED NLRASNLGFQ LYHGVHDGED 240 VKCTPDEGLL LLDLNIRGGY DHRSTADLKM NQHMR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 4e-21 | 26 | 82 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-21 | 26 | 82 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-21 | 26 | 82 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-21 | 26 | 82 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
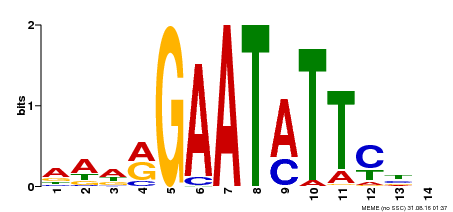
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025808925.1 | 0.0 | myb family transcription factor PHL11-like | ||||
| TrEMBL | A0A2S3GSU5 | 0.0 | A0A2S3GSU5_9POAL; Uncharacterized protein | ||||
| STRING | Si018049m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3940 | 34 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 6e-66 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.A03051.1 |



