 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.A03378.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 326aa MW: 35536.1 Da PI: 4.4698 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 64.6 | 1.4e-20 | 70 | 123 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t+eq++ Le+ Fe+++++ e+++eLA+klgL+ rqV vWFqNrRa++k
Pahal.A03378.1 70 KKRRLTPEQVHLLERSFEEENKLEPERKTELARKLGLQPRQVAVWFQNRRARWK 123
5678*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 132 | 2.3e-42 | 69 | 161 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrl+ eqv+lLE+sFeee+kLeperK+elar+LglqprqvavWFqnrRAR+ktkqlE+d+++Lk+++dal+++++ L +++++Lr+++++
Pahal.A03378.1 69 EKKRRLTPEQVHLLERSFEEENKLEPERKTELARKLGLQPRQVAVWFQNRRARWKTKQLERDFDRLKASFDALRADHDVLLQDNHRLRSQVAS 161
69**************************************************************************************99975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.33E-20 | 63 | 127 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.22 | 65 | 125 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.0E-20 | 68 | 129 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 7.2E-18 | 70 | 123 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 8.85E-21 | 70 | 126 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-22 | 71 | 132 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 2.1E-6 | 96 | 105 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 100 | 123 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 2.1E-6 | 105 | 121 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.8E-17 | 125 | 167 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MESGRLIFNA TGSGAGQMLF LDCGAGGGLF HRGGRPMLGL EEGRGVKRPF FTSPDELLEE 60 EYYDEQLPEK KRRLTPEQVH LLERSFEEEN KLEPERKTEL ARKLGLQPRQ VAVWFQNRRA 120 RWKTKQLERD FDRLKASFDA LRADHDVLLQ DNHRLRSQVA SLTEKLQEKE ATEGGADAEV 180 LPVDVKASLA DDIEEPATEA AAAFEAHQVK SEDRLSTGSG GSAVVDTDAL LCGRFAAAVD 240 SSVESYFPGG EGHYHDCGMG PVNHCAGGIQ SDDDGAGSDE GCSYYPEEAE AAAAAAVAAA 300 FFAGHAHPHA DDDEDASQIG WWMWN* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 117 | 125 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
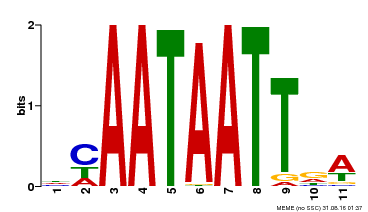
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU961949 | 0.0 | EU961949.1 Zea mays clone 239214 homeodomain leucine zipper protein CPHB-5 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025813024.1 | 0.0 | homeobox-leucine zipper protein HOX16-like isoform X1 | ||||
| Swissprot | Q6YWR4 | 1e-138 | HOX16_ORYSJ; Homeobox-leucine zipper protein HOX16 | ||||
| TrEMBL | A0A2S3GT38 | 0.0 | A0A2S3GT38_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Aa00462.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2609 | 38 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 1e-68 | homeobox 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.A03378.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



