 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.B00059.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 690aa MW: 73949 Da PI: 6.6968 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 89.9 | 2.2e-28 | 202 | 255 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av++L G++kA+Pk+ilelm+v+gLt+e+v+SHLQk+Rl
Pahal.B00059.1 202 KPRVVWSVELHQQFVNAVNHL-GIDKAVPKKILELMNVPGLTRENVASHLQKFRL 255
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 77.5 | 4.9e-26 | 18 | 126 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
vl+vdD+p+ + +l+++l + +y +v+++ +++ al++l+e++ +D+i+ D+ mp+mdG++ll+ e++lp+i+++a + + + + +k G
Pahal.B00059.1 18 VLVVDDDPTCLVVLKRMLLECRY-DVTTCPQATRALTMLRENRrgFDVIISDVHMPDMDGFRLLELVGL-EMDLPVIMMSADSRTDIVMKGIKHG 110
89*********************.***************99999********************87754.558********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ +eel +
Pahal.B00059.1 111 ACDYLIKPVRMEELKN 126
*************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 9.4E-182 | 1 | 672 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 5.0E-42 | 15 | 151 | No hit | No description |
| SuperFamily | SSF52172 | 2.99E-36 | 15 | 140 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 4.7E-30 | 16 | 128 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.196 | 17 | 132 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 5.2E-23 | 18 | 126 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 5.44E-27 | 19 | 131 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-29 | 199 | 260 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.14E-20 | 199 | 259 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.674 | 199 | 258 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 3.1E-25 | 202 | 255 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.8E-7 | 204 | 253 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 690 aa Download sequence Send to blast |
MAAAAEARGG DFPVGMKVLV VDDDPTCLVV LKRMLLECRY DVTTCPQATR ALTMLRENRR 60 GFDVIISDVH MPDMDGFRLL ELVGLEMDLP VIMMSADSRT DIVMKGIKHG ACDYLIKPVR 120 MEELKNIWQH VVRKKFNENK DHEHSGSLDD TDRNRPNNND NEYASSANDG GDGSWKSQKK 180 KREKEEDEAD LESGDPSSTS KKPRVVWSVE LHQQFVNAVN HLGIDKAVPK KILELMNVPG 240 LTRENVASHL QKFRLYLKRI AQHHAGIPHP FVAPASSAKV APLGGLEFQA LAASGQIPPQ 300 ALAALQDELL GRPTSSLALP GRDQSSLRLA AIKGNKPHGE REIAFGQPIY KCQNNAYGAF 360 PQSSPGLGGL PSFGAWPNNK LGMTDSPSAL GNVSNSQNSN LLLHELQQQP DSLLTGTLQN 420 IDVKPSGIVM PSQSLNAFPA SEGISPNQNP LVIPSQSQSF LASIPPSMKH EPLLASSSPS 480 NSLLGGIDMV NQASTSQPLI STHGANLPGL MNRSSNAMPS SGICNFQSGN NPYVVNQNAM 540 GVSSRPPGVL KTESTDSLNR SYGYIGGSTS VDSGLLSSQS KNPQYGLLQN PNDITGSWSP 600 SQDIDSYGNT LGQGHPGSTS SNYQSSNVAL GKLPDQGRGR NHGFVGKGTC IPSRFAVDEV 660 ESPTNNLSHS IGSSADIVNP DIFGFSGQM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 7e-24 | 198 | 261 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
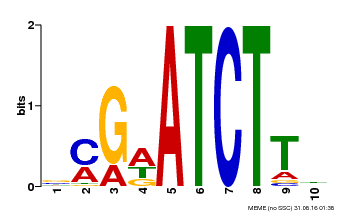
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB060130 | 0.0 | AB060130.1 Zea mays ZmRR8 mRNA for response regulator 8, complete cds. | |||
| GenBank | EU975545 | 0.0 | EU975545.1 Zea mays clone 492147 two-component response regulator ARR1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025794212.1 | 0.0 | two-component response regulator ORR21 | ||||
| Refseq | XP_025794213.1 | 0.0 | two-component response regulator ORR21 | ||||
| Swissprot | A2XE31 | 0.0 | ORR21_ORYSI; Two-component response regulator ORR21 | ||||
| TrEMBL | A0A2S3ISS2 | 0.0 | A0A2S3ISS2_9POAL; Two-component response regulator | ||||
| TrEMBL | A0A2T7CFI5 | 0.0 | A0A2T7CFI5_9POAL; Two-component response regulator | ||||
| STRING | Sb01g042400.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5796 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-114 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.B00059.1 |



