 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.B00461.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 740aa MW: 79649.8 Da PI: 6.7464 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 50.3 | 4.6e-16 | 425 | 463 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
+k+C+CkkskClk+YCeCfaag++Cse C+C++C Nk
Pahal.B00461.2 425 ACKRCSCKKSKCLKLYCECFAAGVYCSEPCSCQGCLNKP 463
589**********************************96 PP
| |||||||
| 2 | TCR | 50.3 | 4.7e-16 | 510 | 549 | 1 | 40 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+gCnCkks+ClkkYCeC++ g+ Cs++C+Ce+CkN+
Pahal.B00461.2 510 RHKRGCNCKKSSCLKKYCECYQGGVGCSSNCRCESCKNTF 549
589***********************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 3.8E-17 | 424 | 465 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.046 | 425 | 550 | IPR005172 | CRC domain |
| Pfam | PF03638 | 8.3E-12 | 427 | 462 | IPR005172 | CRC domain |
| SMART | SM01114 | 3.5E-16 | 510 | 551 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.0E-11 | 512 | 548 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 740 aa Download sequence Send to blast |
MDTPDRAAAQ QPPASRAEDS PVFSFINNLS PIEPLKSAYN ANSLQGYQSI NITSISSIFT 60 SPHDNVHKET RLAKSSLGEI SESEASAGGS KTNKPARSSN AVRLFACTST ITQETHTVTC 120 SDVADPPTGP CDLAQPAQFD NGSPDHNTTP CHGVRSDLKQ DKCRKLDAVQ AVKNTVEKRK 180 CLFSTEIQLL DGGQPVNDSD DVLGCEWSDL ISTTSGDSEI SGRAHPSASG QVYYQELVMG 240 EDQTENPQIF QDGQQTISTE EIQDNIYEPN GCIPLDYKVE SQQRGIRRRC LVFEAAGFSN 300 TVVQKETVES LSVSTCKGKS HVQTQPRGLR GIGLHLNALA LTPKGKMASQ DHMASGLRPS 360 LAEKDVHGKF LSAGENFPNS GGELLEFPMD DCSAGGFPVN DHVSSQSVSP QKKRRKTDNG 420 DDGEACKRCS CKKSKCLKLY CECFAAGVYC SEPCSCQGCL NKPIHEEIVL STRKQIEFRN 480 PLAFAPKVIR MSDAGPETGE DPNSTPASAR HKRGCNCKKS SCLKKYCECY QGGVGCSSNC 540 RCESCKNTFG RRDADSELIE EIKQEGEQTE NCGKEKENDQ QKANVQNEDH PLLDLVPITP 600 PFDLSSSLLK LPNFSSAKPP RPSKARSGNY RSSASKATAT LQSCKSSKVA GSAIDEGMPD 660 ILKEADSPNN CVKTTSPNGK RVSPPHNALS ISPNRKGGRK LILKSIPSFP SLMGESNSGS 720 AMNDTDNTFN ASPLVLGPS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 3e-15 | 429 | 547 | 14 | 120 | Protein lin-54 homolog |
| 5fd3_B | 3e-15 | 429 | 547 | 14 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 411 | 415 | KKRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
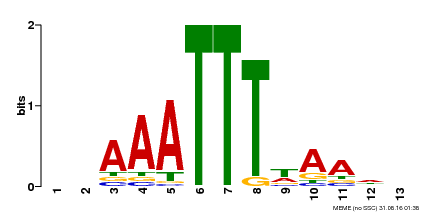
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025799594.1 | 0.0 | protein tesmin/TSO1-like CXC 2 | ||||
| TrEMBL | A0A2S3GW28 | 0.0 | A0A2S3GW28_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Bb00411.1.p | 0.0 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 3e-87 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.B00461.2 |



