 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.B03330.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 497aa MW: 55168.1 Da PI: 7.3172 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.8 | 3.5e-11 | 188 | 231 | 1 | 41 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHH...HHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLee...kvkeLeae 41
e++ k rr+++NReAAr+sR+RKka+i++Le + ++Le+e
Pahal.B03330.1 188 ERDPKTLRRLAQNREAARKSRLRKKAYIQQLESsriRLAQLEQE 231
6777999************************9633355555555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.8E-8 | 184 | 236 | No hit | No description |
| SMART | SM00338 | 5.6E-8 | 188 | 253 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.3E-8 | 189 | 231 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.082 | 190 | 232 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.46E-7 | 193 | 234 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 195 | 210 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.3E-28 | 288 | 362 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 497 aa Download sequence Send to blast |
MASGGSSVRE QQQEMNISFG MMNHHHGHHH QQPPSSSSSS SMHAAAASFM SGKEASGAYD 60 HLGELDQALF MYLDHGSGHG ATHQEQRQTL NIFPSQPMHV EPSPKGEISL VLSPAPVGSK 120 QPRSPDHHHH QQAAMEELAG SRRLQQEHHH LQHQPFPAAG AEPAAPGMIK DVKPLTKKDH 180 RRGTSTSERD PKTLRRLAQN REAARKSRLR KKAYIQQLES SRIRLAQLEQ ELHTARAQGV 240 FFPNSGLLAD QGVAGKGVPI GGIDGLSSEA AMFDVEYGRW QEEHYRLMYE LRAALQQHLP 300 EGELQMYVES CLAHHDEMMG IKEGAIKGDV FHLISGVWRS PAERCFLWLG GFRPSEVIKM 360 LLSHVEPLTE QQIVGVYGLQ QSALETEEAL SQGLDALYQS LSDTVVSDAL SCPSNVANYM 420 GQMAAAMNKL STLEGFVRQA ESLRQQTLHR LHQILTTRQM ARSLLAVSDY FHRLRTLSSL 480 WVTRPRAPQE QQQGHS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to as1-like elements (5'-TGACGTAAgggaTGACGCA-3') in promoters of target genes. Regulates transcription in response to plant signaling molecules salicylic acid (SA), methyl jasmonate (MJ) and auxin (2,4D) only in leaves. Prevents lateral branching and may repress defense signaling. {ECO:0000269|PubMed:12777042}. | |||||
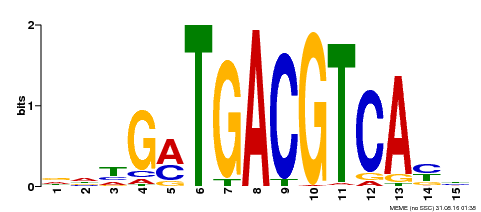
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00491 | DAP | Transfer from AT5G06839 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU966928 | 0.0 | EU966928.1 Zea mays clone 298194 DNA binding protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025800643.1 | 0.0 | transcription factor TGA2.1-like | ||||
| Swissprot | Q52MZ2 | 1e-172 | TGA10_TOBAC; bZIP transcription factor TGA10 | ||||
| TrEMBL | A0A2S3H0U7 | 0.0 | A0A2S3H0U7_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G006578_P03 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7876 | 35 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06839.3 | 1e-146 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.B03330.1 |



