 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.B03967.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 884aa MW: 99347.4 Da PI: 9.0438 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 164.4 | 2e-51 | 4 | 117 | 4 | 117 |
CG-1 4 ekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrc 98
+rw +++ei+a+L+n++++++++++ ++p sg+++Ly+rk+vr+frkDG++wkkkkdgktv+E+hekLK+g+ e +++yYa++ee+p+f rrc
Pahal.B03967.2 4 AGTRWFRPNEIYAVLANHARFKVHAQPVDKPASGTVVLYDRKVVRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEEKVHVYYARGEEDPNFFRRC 98
478******************************************************************************************** PP
CG-1 99 ywlLeeelekivlvhylev 117
ywlL++ele+ivlvhy+++
Pahal.B03967.2 99 YWLLDKELERIVLVHYRQT 117
****************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 72.248 | 1 | 123 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.5E-60 | 1 | 118 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.1E-44 | 4 | 116 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 6.44E-11 | 350 | 436 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 6.22E-18 | 495 | 639 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.17E-16 | 527 | 635 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.0E-18 | 534 | 642 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.298 | 543 | 647 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.6E-8 | 549 | 639 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.808 | 576 | 608 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 6.3E-6 | 576 | 605 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 560 | 615 | 644 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 9.82E-11 | 690 | 792 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.24 | 741 | 763 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 742 | 771 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0055 | 744 | 762 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0019 | 764 | 786 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.53 | 765 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 7.0E-4 | 766 | 786 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 7.3 | 845 | 867 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.249 | 847 | 875 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.12 | 848 | 867 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 884 aa Download sequence Send to blast |
MAEAGTRWFR PNEIYAVLAN HARFKVHAQP VDKPASGTVV LYDRKVVRNF RKDGHNWKKK 60 KDGKTVQEAH EKLKIGNEEK VHVYYARGEE DPNFFRRCYW LLDKELERIV LVHYRQTSEE 120 NAIPPPHIEA EVAEVPPINI IHYTSPIIST DSASARTELS SCAAAAAPEE INSHGGGAIS 180 CETDDHDSSL ESFWADLLES SMKNDTSVRG GSLTPNQQTN DGMRDSGNSI LNTNARSNAI 240 FSSPANVVSE AYAANPGLNQ VSESYYGALK HQVNQSPSLL TSDLDSQSKP HANSIIKTLV 300 DGNMPSDVPA RQNSLGLWKY LDDDITCLVN NPSSAIPITR PVIDEMPFHI IEISSEWAYC 360 TDDTKVLVVG YFLDNYKHLS GANLYCVIGE QCVTADIVQT GVYRFMARPH VPGRVNLYLT 420 LDGKTPISEV LSFEYRRMLG SSDDDEPKKS KLQMQMRLAR LLFSTSQKKI PPKFLVEGSR 480 VSNLLSASTE KEWMNLFKYV TDSKGTNIPA TESLLELVLR NKLQEWLVEK IIEGHKSTDR 540 DDLGQGPIHL CSFLGYTWAI RLFSLSGFSL DFRDSSGWTA LHWAAYYGRE KMVAALLSAG 600 ANPSLVTDPT HDDPGGHTAA DIAARQGFDG LAAYLAEKGL TAHFEAMSLS KDKKSTSRTQ 660 SIKQISKEFE NLTEQELCLR ESLAAYRNAA DAASNIQAAL RERTLKLQTK AIQSANPEIE 720 AATIVAAMRI QHAFRNYNRK KMMRAAARIQ SHFRTWQMRR NFMNMRRQAI KIQAAYRGHQ 780 VRRQYRKVIW SVGVVEKAIL RWRKKRKGLR GIATGMPVAM STDTEAASTA EEDYYQVGRQ 840 QAENRFNRSV VRVQALFRSH RAQQEYRRMK VAHEEAKVEF SQN* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
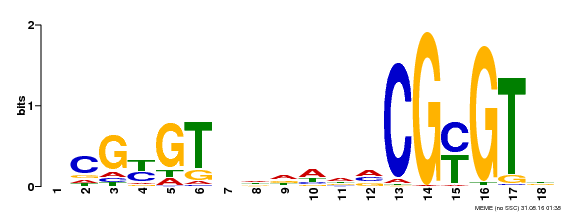
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025800750.1 | 0.0 | calmodulin-binding transcription activator CBT isoform X1 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A2S3H2F3 | 0.0 | A0A2S3H2F3_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ba00948.1.p | 0.0 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.B03967.2 |



